KN motif and ankyrin repeat domains 4
ZFIN 
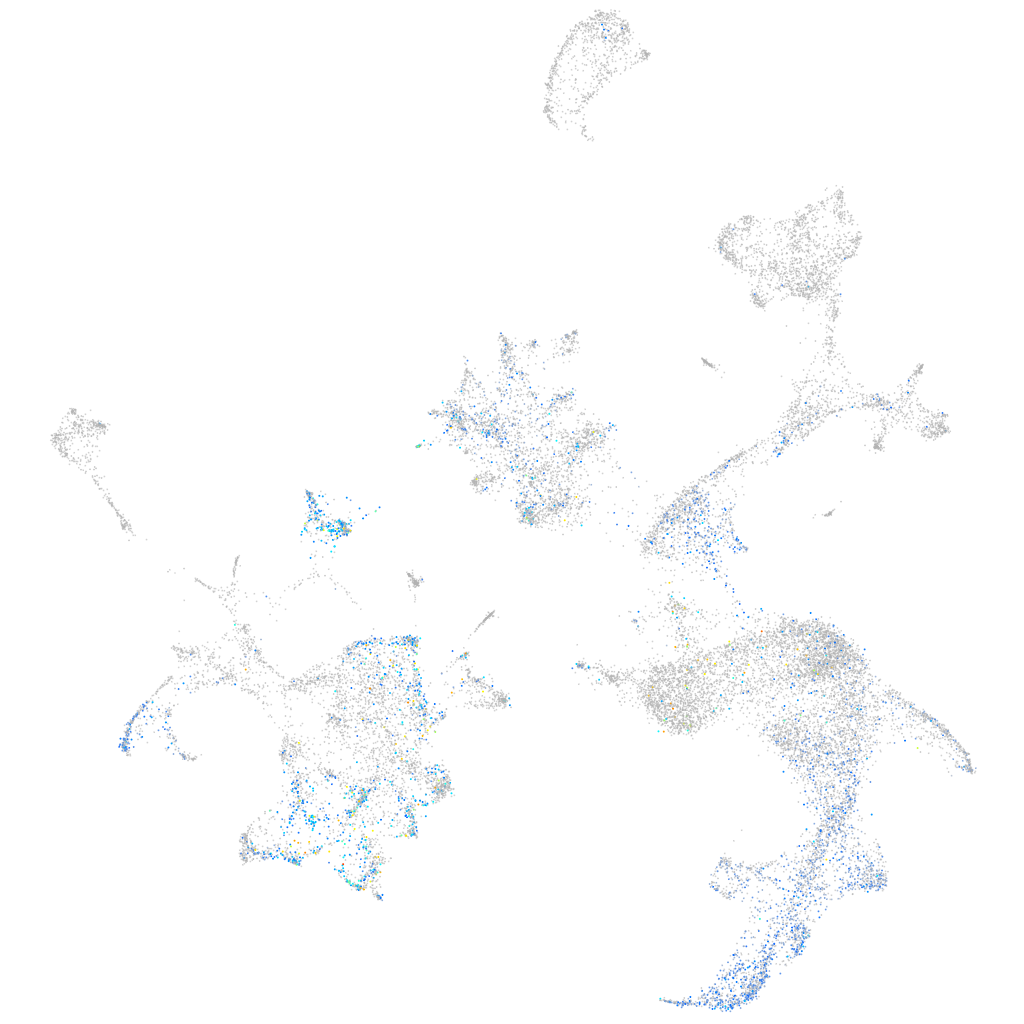
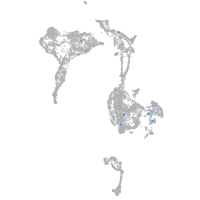
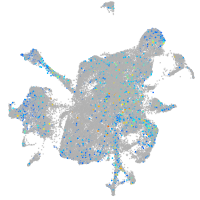
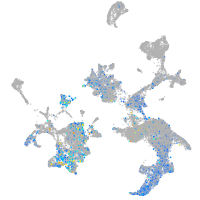
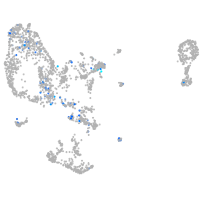
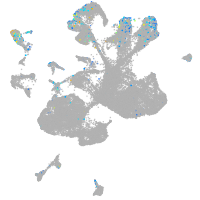
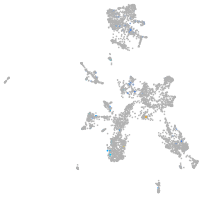
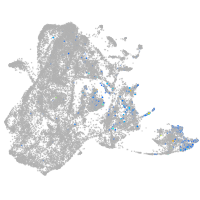
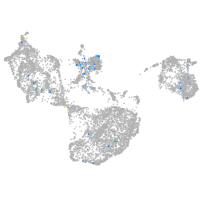
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| krt94 | 0.209 | coro1a | -0.115 |
| cav1 | 0.192 | laptm5 | -0.107 |
| col4a1 | 0.186 | gpx1a | -0.104 |
| limch1b | 0.184 | rac2 | -0.103 |
| ptprb | 0.177 | pfn1 | -0.100 |
| cavin2b | 0.174 | fcer1gl | -0.095 |
| ehd2b | 0.173 | wasb | -0.094 |
| fam174b | 0.172 | arpc1b | -0.090 |
| cavin1b | 0.172 | ctss2.1 | -0.090 |
| krt8 | 0.169 | grap2b | -0.089 |
| eppk1 | 0.166 | spi1b | -0.089 |
| edn1 | 0.165 | ptpn6 | -0.088 |
| igfbp7 | 0.165 | si:ch211-102c2.4 | -0.087 |
| gstt1b | 0.163 | fam49ba | -0.083 |
| clec14a | 0.163 | zgc:64051 | -0.081 |
| anxa13 | 0.162 | cebpb | -0.080 |
| klf3 | 0.161 | glipr1a | -0.080 |
| tspan4b | 0.161 | hcls1 | -0.080 |
| si:ch211-145b13.6 | 0.159 | ccr9a | -0.080 |
| klf2a | 0.158 | FERMT3 (1 of many) | -0.079 |
| krt18a.1 | 0.157 | grap2a | -0.078 |
| shroom4 | 0.156 | CU499330.1 | -0.078 |
| col4a2 | 0.156 | rgs13 | -0.077 |
| myl9a | 0.156 | si:dkey-102g19.3 | -0.076 |
| fabp11a | 0.155 | cxcr3.2 | -0.075 |
| cast | 0.155 | lcp1 | -0.075 |
| myo1ca | 0.155 | cotl1 | -0.074 |
| cdh5 | 0.155 | itgae.2 | -0.074 |
| acvrl1 | 0.155 | lgals2a | -0.074 |
| plecb | 0.153 | il1b | -0.074 |
| fstl1b | 0.153 | si:dkey-262k9.4 | -0.073 |
| si:dkeyp-97a10.2 | 0.151 | ncf1 | -0.073 |
| cfd | 0.151 | ptprc | -0.073 |
| ahnak | 0.150 | plek | -0.072 |
| pecam1 | 0.150 | LOC100536213 | -0.072 |