kalirin RhoGEF kinase b
ZFIN 
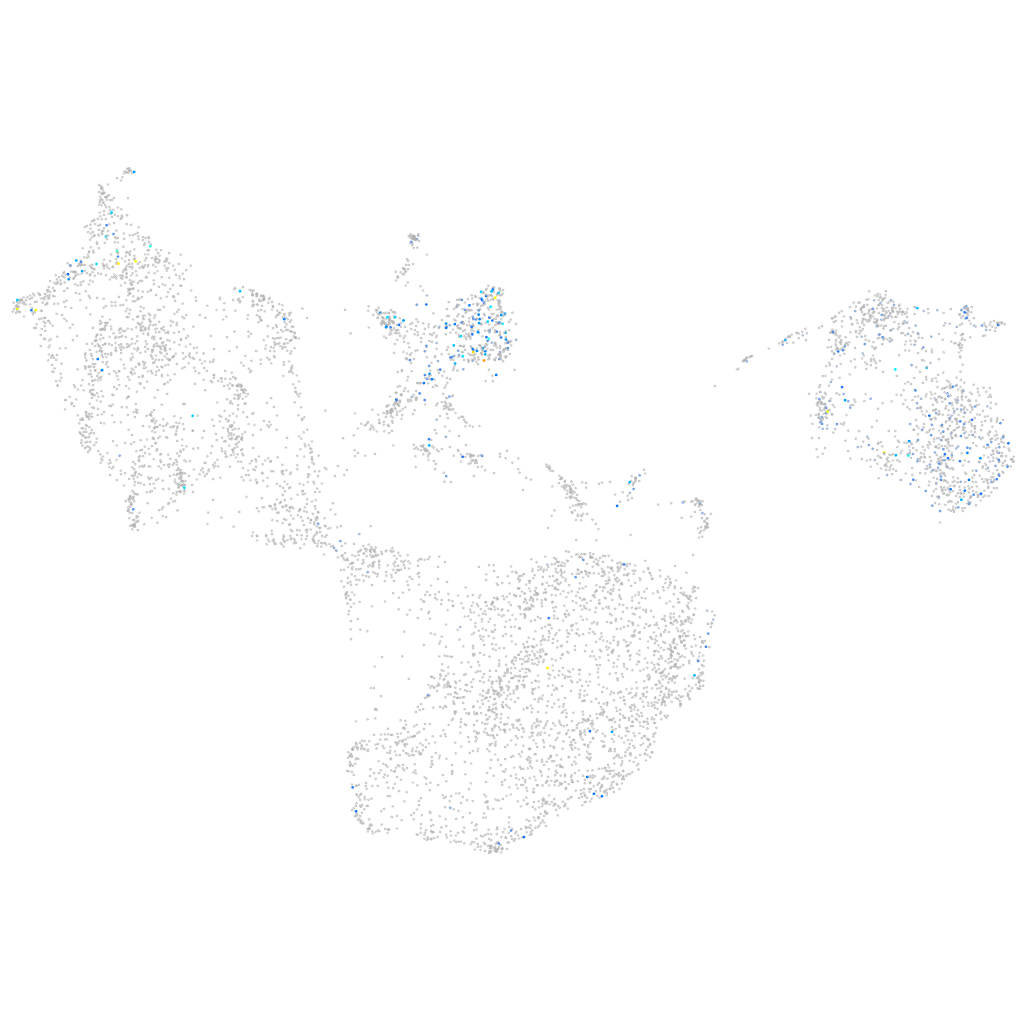
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gap43 | 0.201 | mdka | -0.070 |
| myt1la | 0.200 | sparc | -0.066 |
| bsnb | 0.200 | fkbp7 | -0.066 |
| gng3 | 0.199 | ccng1 | -0.064 |
| gpm6ab | 0.196 | ppib | -0.058 |
| ywhag2 | 0.195 | fkbp14 | -0.057 |
| stmn1b | 0.191 | fkbp9 | -0.056 |
| stmn2a | 0.190 | frem3 | -0.054 |
| phactr3a | 0.185 | hoxa13a | -0.053 |
| gpm6aa | 0.184 | zgc:101810 | -0.052 |
| adgrb3 | 0.183 | crabp1b | -0.051 |
| stx1b | 0.182 | fam49ba | -0.051 |
| rtn1b | 0.182 | id1 | -0.051 |
| serp2 | 0.181 | col1a1a | -0.050 |
| dpysl3 | 0.181 | actn1 | -0.049 |
| rtn1a | 0.180 | fkbp10b | -0.049 |
| sncb | 0.178 | sri | -0.049 |
| stxbp1a | 0.176 | hoxc13a | -0.048 |
| si:dkeyp-117h8.2 | 0.173 | pak2a | -0.048 |
| zgc:153426 | 0.173 | cldn1 | -0.048 |
| tuba1c | 0.172 | hcls1 | -0.048 |
| si:ch73-60h1.1 | 0.171 | crtap | -0.047 |
| si:ch211-257p13.3 | 0.169 | fat2 | -0.047 |
| mllt11 | 0.169 | plod1a | -0.046 |
| tmsb | 0.169 | arhgef25b | -0.046 |
| elavl3 | 0.167 | zgc:171775 | -0.046 |
| ppp1r14ba | 0.167 | rpl29 | -0.045 |
| nova2 | 0.164 | si:dkey-52l18.4 | -0.045 |
| zgc:65894 | 0.164 | vcana | -0.045 |
| CR786577.1 | 0.163 | rplp1 | -0.045 |
| atp6v0cb | 0.163 | egfl6 | -0.045 |
| mdkb | 0.162 | pdlim1 | -0.045 |
| mturn | 0.162 | pxna | -0.045 |
| csdc2a | 0.162 | GCA | -0.044 |
| si:ch73-290k24.6 | 0.162 | and1 | -0.044 |