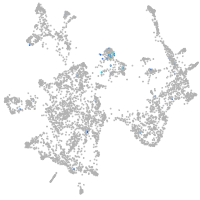
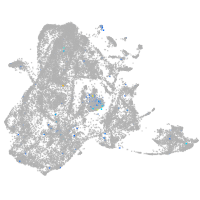
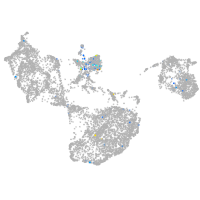
junctophilin 3
ZFIN 
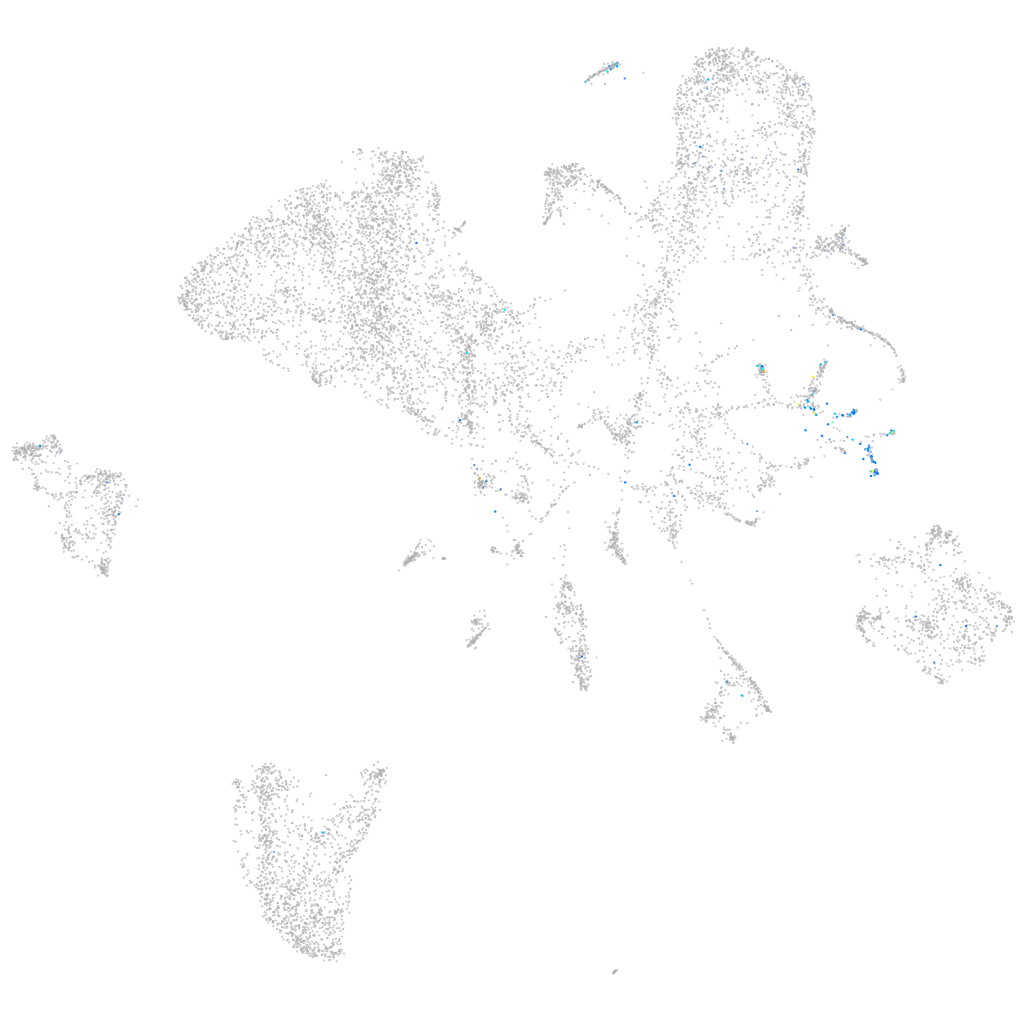
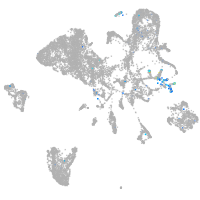
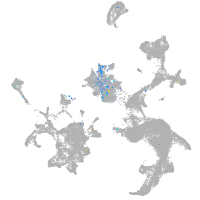
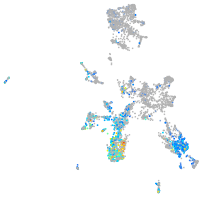
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rims2a | 0.370 | ahcy | -0.115 |
| scg3 | 0.354 | gapdh | -0.102 |
| pax6b | 0.337 | rps20 | -0.094 |
| neurod1 | 0.327 | gamt | -0.094 |
| scgn | 0.326 | hdlbpa | -0.090 |
| si:dkey-153k10.9 | 0.324 | aldob | -0.087 |
| syt1a | 0.322 | gatm | -0.086 |
| pcsk1nl | 0.322 | gstp1 | -0.083 |
| pcsk1 | 0.320 | fbp1b | -0.082 |
| vat1 | 0.320 | nupr1b | -0.082 |
| camk1da | 0.312 | fabp3 | -0.081 |
| scg2b | 0.310 | bhmt | -0.081 |
| dkk3b | 0.306 | rpl6 | -0.081 |
| SLC5A10 | 0.304 | gstr | -0.080 |
| rprmb | 0.303 | gstt1a | -0.079 |
| scg2a | 0.301 | cebpd | -0.078 |
| c2cd4a | 0.286 | mat1a | -0.077 |
| slc30a2 | 0.282 | aqp12 | -0.076 |
| isl1 | 0.282 | cx32.3 | -0.076 |
| aldocb | 0.277 | apoa4b.1 | -0.076 |
| ptn | 0.275 | aldh6a1 | -0.075 |
| kcnj11 | 0.275 | apoa1b | -0.075 |
| cacna1ea | 0.274 | gnmt | -0.074 |
| insm1a | 0.273 | eno3 | -0.074 |
| cntnap2a | 0.271 | hspe1 | -0.073 |
| cpe | 0.267 | si:dkey-16p21.8 | -0.073 |
| mir7a-1 | 0.267 | apoc2 | -0.072 |
| pcdh20 | 0.265 | cebpb | -0.072 |
| tspan7b | 0.264 | aldh1l1 | -0.071 |
| nkx2.2a | 0.261 | aldh7a1 | -0.071 |
| ndufa4l2a | 0.259 | shmt1 | -0.070 |
| foxo1a | 0.257 | agxtb | -0.070 |
| sgip1b | 0.254 | agxta | -0.070 |
| vamp2 | 0.254 | dhdhl | -0.069 |
| dgkh | 0.254 | tktb | -0.069 |