intersectin 1 (SH3 domain protein)
ZFIN 
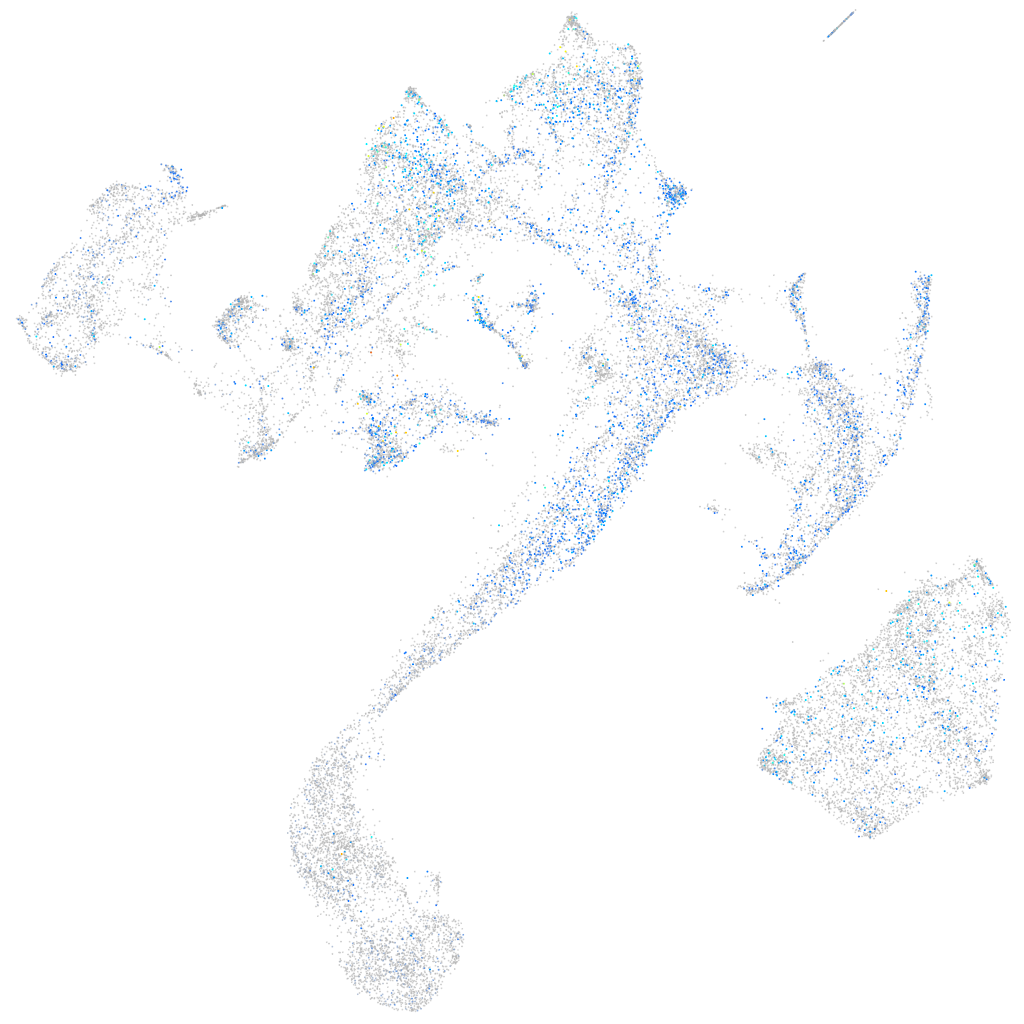
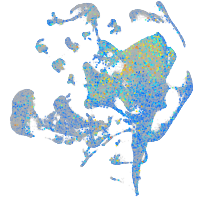
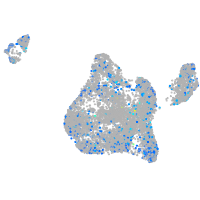
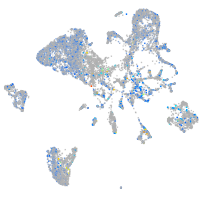
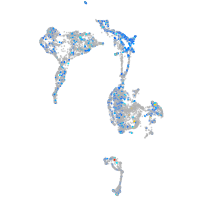
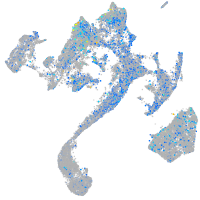
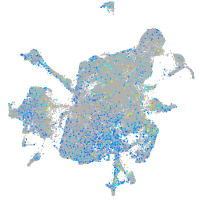
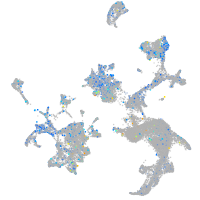
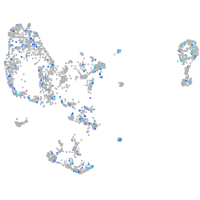
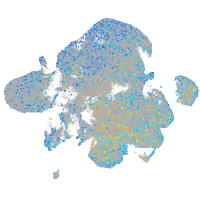
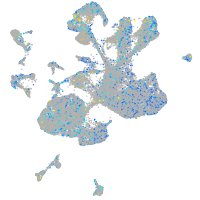
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mfap2 | 0.134 | ckma | -0.110 |
| mdka | 0.132 | ckmb | -0.105 |
| fstl1b | 0.132 | tnnt3a | -0.105 |
| marcksl1a | 0.129 | si:ch73-367p23.2 | -0.105 |
| actb2 | 0.127 | gapdh | -0.103 |
| cd81a | 0.127 | actn3a | -0.103 |
| CR383676.1 | 0.127 | tnnc2 | -0.102 |
| emp2 | 0.126 | myom1a | -0.101 |
| ssr3 | 0.124 | ldb3b | -0.101 |
| col5a1 | 0.121 | myoz1b | -0.100 |
| ppib | 0.119 | smyd1a | -0.100 |
| col5a2a | 0.116 | mylz3 | -0.100 |
| sparc | 0.114 | ak1 | -0.099 |
| hnrnpa0l | 0.114 | neb | -0.099 |
| h3f3a | 0.113 | atp2a1 | -0.098 |
| actb1 | 0.112 | si:ch211-266g18.10 | -0.097 |
| tpm4a | 0.112 | XLOC-001975 | -0.097 |
| fbn2b | 0.112 | tmod4 | -0.097 |
| foxp4 | 0.112 | mylpfb | -0.097 |
| tmem258 | 0.111 | XLOC-025819 | -0.097 |
| sdc4 | 0.110 | tnnt3b | -0.096 |
| colec12 | 0.110 | pgam2 | -0.096 |
| col1a2 | 0.110 | tnni2a.4 | -0.096 |
| hsp90ab1 | 0.109 | ank1a | -0.095 |
| fstl1a | 0.108 | prr33 | -0.095 |
| ssr4 | 0.108 | hhatla | -0.094 |
| zgc:153867 | 0.107 | XLOC-005350 | -0.094 |
| reck | 0.107 | myom2a | -0.093 |
| ssr2 | 0.106 | XLOC-006515 | -0.093 |
| col1a1a | 0.106 | prx | -0.093 |
| col1a1b | 0.106 | dhrs7cb | -0.092 |
| fkbp9 | 0.106 | cavin4b | -0.092 |
| zgc:92744 | 0.105 | si:ch211-255p10.3 | -0.092 |
| lrp1ab | 0.105 | pvalb1 | -0.092 |
| h3f3c | 0.105 | slc25a4 | -0.091 |