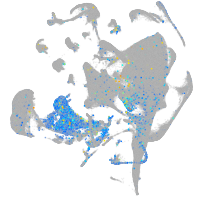
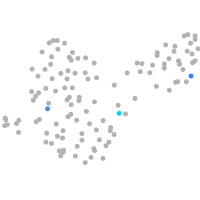
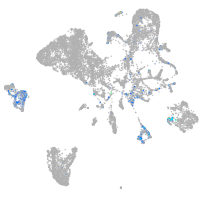
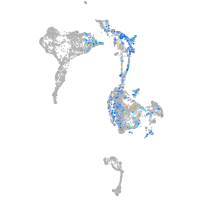
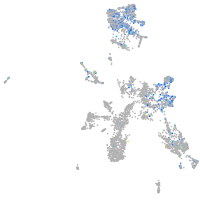
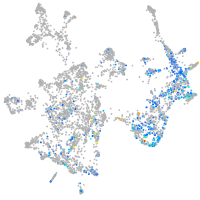
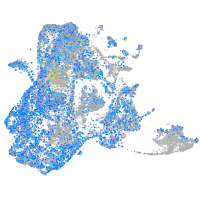
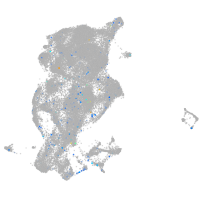
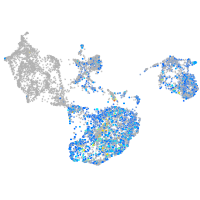
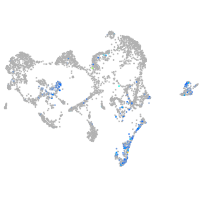
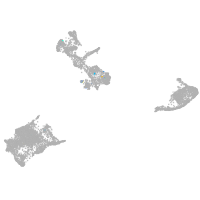
"integrin, alpha 3b"
ZFIN 
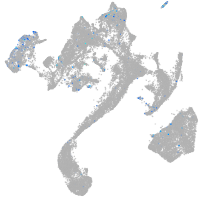
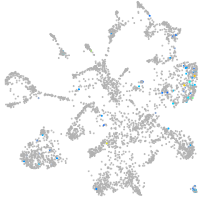
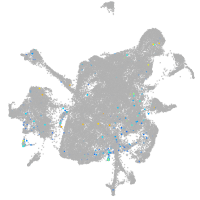
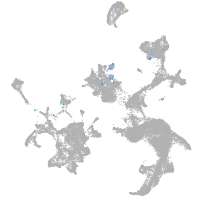
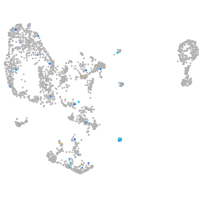
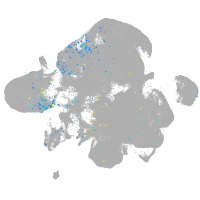
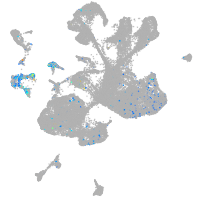
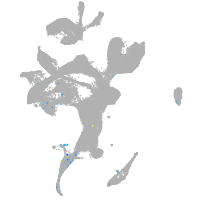
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| nphs2 | 0.265 | bhmt | -0.040 |
| si:ch73-205h11.1 | 0.228 | pmp22a | -0.039 |
| ptprh | 0.227 | krt18b | -0.034 |
| ptprq | 0.211 | LO018188.1 | -0.033 |
| nphs1 | 0.184 | fibina | -0.033 |
| itgb6 | 0.171 | six1a | -0.030 |
| naalad2 | 0.157 | snai1a | -0.029 |
| si:dkey-1c11.1 | 0.149 | CU929237.1 | -0.028 |
| fam49ba | 0.146 | prrx1b | -0.026 |
| lmx1a | 0.143 | cdh11 | -0.026 |
| il2rb | 0.141 | rarga | -0.026 |
| bcam | 0.135 | fkbp7 | -0.025 |
| lama5 | 0.131 | col1a2 | -0.025 |
| col4a4 | 0.116 | fgfbp2b | -0.025 |
| podxl | 0.112 | twist1b | -0.025 |
| iqgap2 | 0.111 | twist1a | -0.024 |
| wt1b | 0.108 | adh5 | -0.024 |
| cldn5a | 0.106 | tuba8l3 | -0.024 |
| col4a3 | 0.105 | rps9 | -0.024 |
| sh3d19 | 0.104 | dlx5a | -0.024 |
| XLOC-011909 | 0.099 | gsc | -0.024 |
| cnksr1 | 0.099 | dcn | -0.024 |
| vwa2 | 0.096 | KCTD15 | -0.024 |
| lrmp | 0.095 | loxl1 | -0.023 |
| vegfba | 0.095 | rps20 | -0.023 |
| slc9a3r1a | 0.093 | mxra8b | -0.023 |
| b3gat2 | 0.092 | lum | -0.022 |
| sema7a | 0.092 | sod1 | -0.022 |
| magi2a | 0.092 | fli1a | -0.022 |
| hepacama | 0.092 | lpar1 | -0.021 |
| map3k15 | 0.091 | col9a1a | -0.021 |
| dachc | 0.091 | cxcl12a | -0.021 |
| ccdc162 | 0.090 | ldhba | -0.021 |
| dag1 | 0.085 | ccl25b | -0.021 |
| spata13 | 0.084 | VIT | -0.021 |