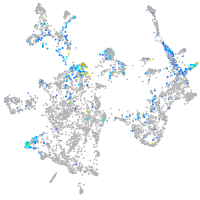
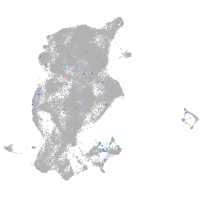
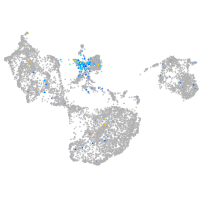
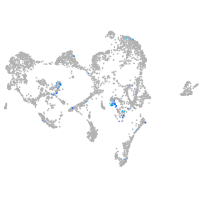
ISL LIM homeobox 1
ZFIN 
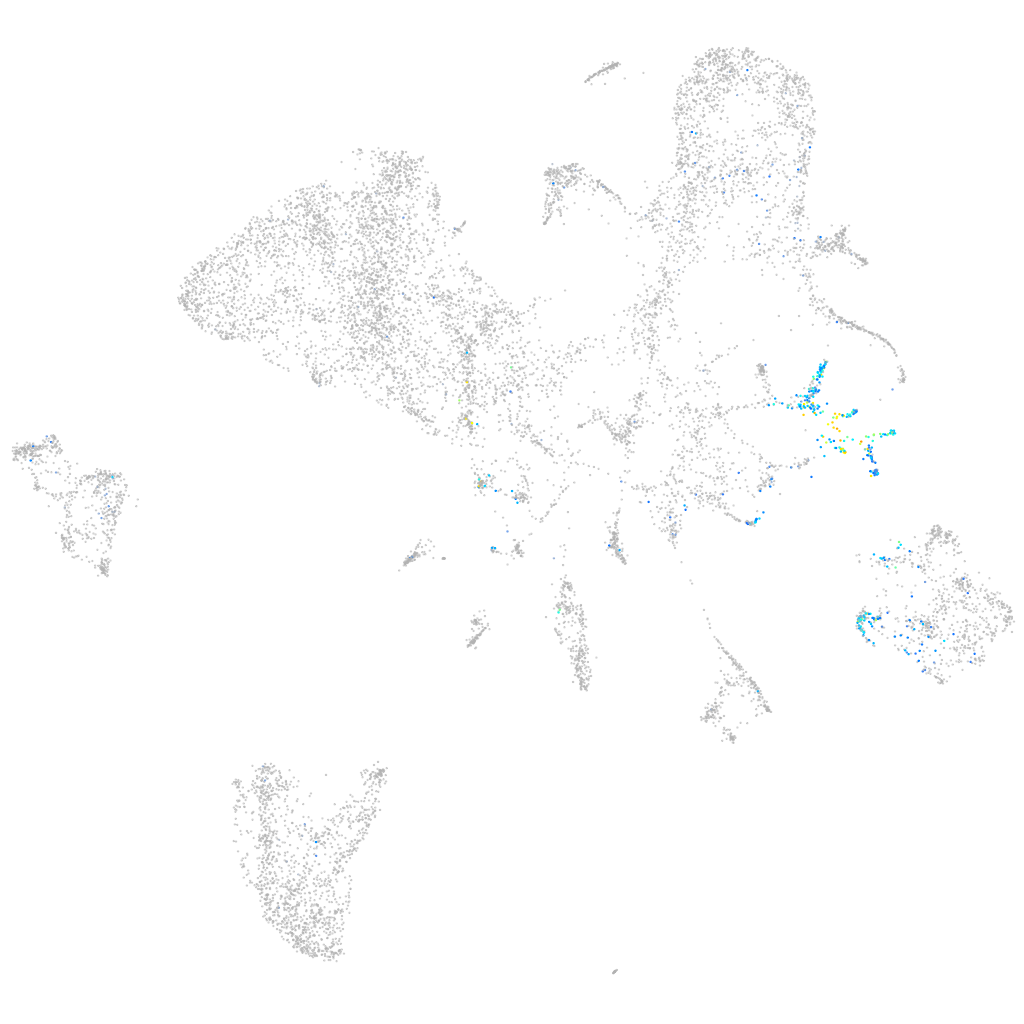
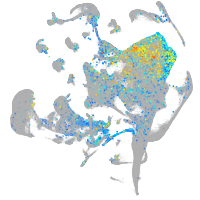
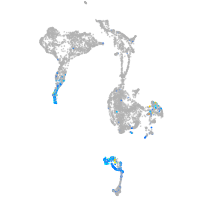
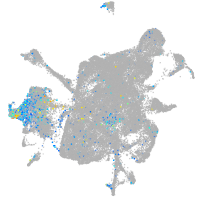
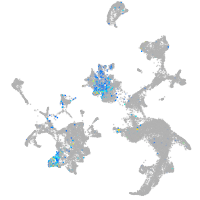
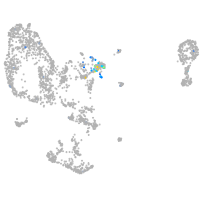
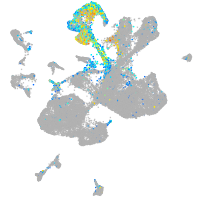
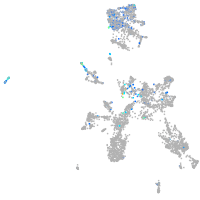
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scgn | 0.633 | ahcy | -0.215 |
| kcnj11 | 0.538 | gapdh | -0.202 |
| neurod1 | 0.535 | aldob | -0.194 |
| rprmb | 0.527 | gamt | -0.184 |
| pax6b | 0.523 | eno3 | -0.168 |
| si:dkey-153k10.9 | 0.521 | gstp1 | -0.167 |
| insm1a | 0.502 | gatm | -0.163 |
| scg3 | 0.498 | eef1da | -0.163 |
| arxa | 0.485 | hdlbpa | -0.159 |
| syt1a | 0.438 | aldh6a1 | -0.159 |
| pcsk2 | 0.437 | fbp1b | -0.155 |
| dkk3b | 0.429 | bhmt | -0.155 |
| scg2a | 0.428 | glud1b | -0.153 |
| pcsk1nl | 0.420 | fabp3 | -0.151 |
| mir7a-1 | 0.416 | gstt1a | -0.148 |
| gcgb | 0.411 | mat1a | -0.148 |
| LOC100537384 | 0.403 | aldh7a1 | -0.148 |
| tspan7b | 0.402 | nupr1b | -0.147 |
| camk1da | 0.398 | apoa1b | -0.146 |
| ndufa4l2a | 0.392 | cx32.3 | -0.143 |
| myt1b | 0.389 | apoa4b.1 | -0.142 |
| abhd15a | 0.384 | gstr | -0.142 |
| nkx2.2a | 0.384 | prdx6 | -0.140 |
| vat1 | 0.381 | apoc2 | -0.139 |
| slc35g2b | 0.377 | ppa1b | -0.139 |
| gcga | 0.377 | ckba | -0.138 |
| ghrl | 0.377 | cebpd | -0.136 |
| sst2 | 0.374 | gnmt | -0.135 |
| rasd4 | 0.373 | afp4 | -0.134 |
| SLC5A10 | 0.373 | scp2a | -0.134 |
| cacna1c | 0.372 | suclg1 | -0.133 |
| fam49bb | 0.371 | agxta | -0.133 |
| CR774179.5 | 0.370 | suclg2 | -0.132 |
| slc1a4 | 0.369 | gcshb | -0.131 |
| c2cd4a | 0.366 | aqp12 | -0.131 |