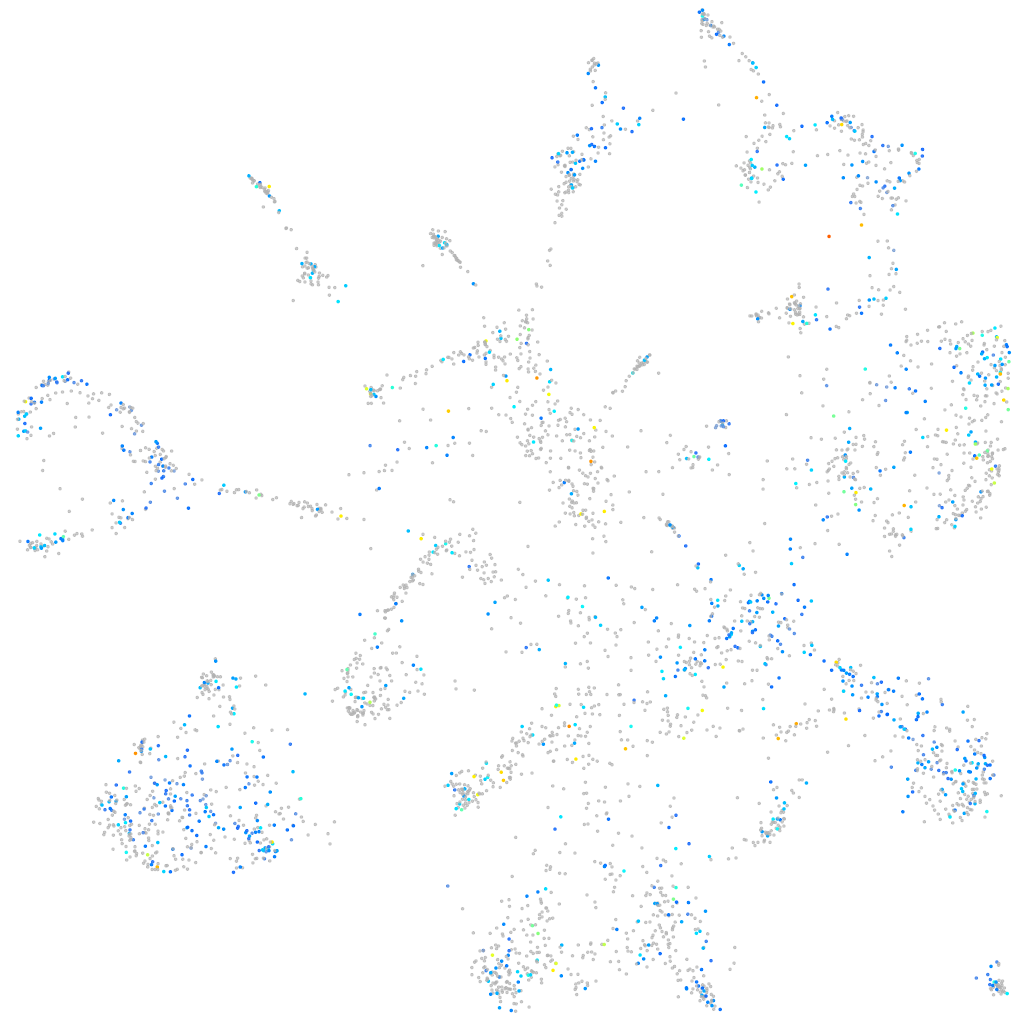
importin 7
ZFIN 
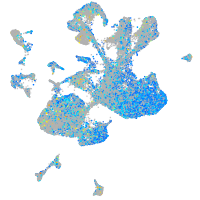
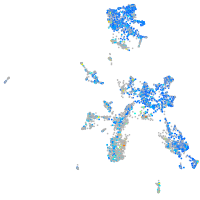
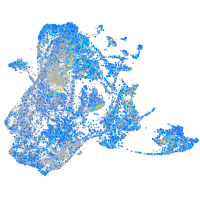
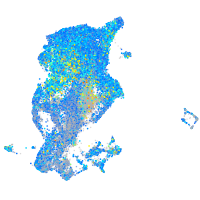
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| pcna | 0.151 | tmsb4x | -0.084 |
| mcm6 | 0.142 | pvalb1 | -0.067 |
| lig1 | 0.139 | mfap2 | -0.065 |
| stmn1a | 0.131 | mylpfa | -0.064 |
| slbp | 0.126 | cotl1 | -0.063 |
| rrm2 | 0.125 | krt4 | -0.062 |
| cdca7a | 0.124 | actc1b | -0.061 |
| dut | 0.121 | pvalb2 | -0.059 |
| rbbp4 | 0.120 | ppt1 | -0.059 |
| chaf1a | 0.120 | sept10 | -0.057 |
| msh6 | 0.120 | zgc:153867 | -0.057 |
| hells | 0.119 | sparc | -0.054 |
| rrm1 | 0.119 | calm2a | -0.053 |
| baz1b | 0.118 | si:dkeyp-75h12.5 | -0.052 |
| CABZ01005379.1 | 0.118 | tpma | -0.052 |
| fbl | 0.115 | gngt2b | -0.051 |
| hmga1a | 0.113 | gng10 | -0.051 |
| mcm2 | 0.113 | myl9a | -0.050 |
| kpnb1 | 0.111 | tpm1 | -0.050 |
| selenoh | 0.111 | ckba | -0.050 |
| npm1a | 0.110 | apoa2 | -0.048 |
| zgc:110540 | 0.110 | c1qtnf1 | -0.048 |
| mcm5 | 0.109 | igfbp7 | -0.047 |
| dhfr | 0.108 | enpp6 | -0.047 |
| ncl | 0.107 | si:ch211-226m16.2 | -0.046 |
| ssrp1a | 0.105 | mcl1a | -0.046 |
| ppm1g | 0.104 | zgc:123103 | -0.046 |
| asf1ba | 0.104 | esama | -0.045 |
| polr2h | 0.103 | si:ch73-335l21.4 | -0.045 |
| si:ch1073-228h2.2 | 0.103 | trappc4 | -0.045 |
| tk1 | 0.103 | si:ch211-195b11.3 | -0.045 |
| parp1 | 0.102 | tram2 | -0.044 |
| dnmt1 | 0.102 | zgc:174863 | -0.044 |
| nop58 | 0.102 | myl10 | -0.044 |
| seta | 0.101 | prss35 | -0.044 |