immunoglobulin superfamily member 11
ZFIN 
Other cell groups


all cells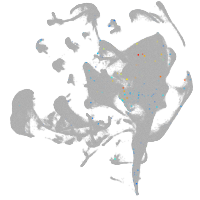 |
blastomeres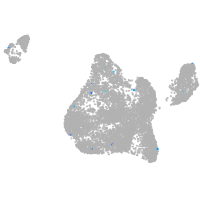 |
PGCs |
endoderm |
axial mesoderm |
muscle 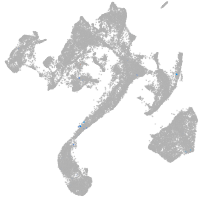 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural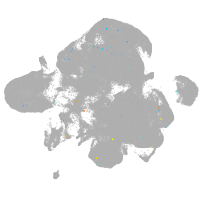 |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hepacama | 0.034 | krt4 | -0.012 |
| si:ch211-66e2.5 | 0.034 | wu:fb18f06 | -0.012 |
| slc6a11b | 0.033 | cyt1 | -0.011 |
| atp1a1b | 0.031 | cyt1l | -0.011 |
| qki2 | 0.031 | icn | -0.011 |
| si:dkey-245n4.2 | 0.030 | krt5 | -0.011 |
| slc1a3b | 0.030 | pycard | -0.011 |
| gpr37l1b | 0.029 | tmsb1 | -0.011 |
| sept8b | 0.029 | tuba8l2 | -0.011 |
| slc1a2b | 0.029 | zgc:153284 | -0.011 |
| gpm6bb | 0.028 | zgc:193505 | -0.011 |
| atp1b4 | 0.027 | agr1 | -0.010 |
| si:ch1073-303k11.2 | 0.027 | anxa1a | -0.010 |
| cldn7a | 0.026 | anxa1c | -0.010 |
| fabp7a | 0.026 | cfl1l | -0.010 |
| heyl | 0.026 | ckap4 | -0.010 |
| ptn | 0.025 | cldne | -0.010 |
| ptprz1b | 0.024 | epcam | -0.010 |
| si:ch73-265d7.2 | 0.024 | icn2 | -0.010 |
| sox10 | 0.024 | krtt1c19e | -0.010 |
| zgc:165461 | 0.024 | lye | -0.010 |
| fam181b | 0.023 | mgst1.2 | -0.010 |
| gfap | 0.023 | mid1ip1a | -0.010 |
| qkib | 0.023 | s100a10b | -0.010 |
| slc4a4a | 0.023 | si:ch211-195b11.3 | -0.010 |
| BX510923.1 | 0.022 | si:dkey-87o1.2 | -0.010 |
| CR848047.1 | 0.022 | zgc:175088 | -0.010 |
| efhd1 | 0.022 | abcb5 | -0.009 |
| mlc1 | 0.022 | aep1 | -0.009 |
| cdc42ep1b | 0.022 | anxa1b | -0.009 |
| FAM107A (1 of many) | 0.022 | anxa3a | -0.009 |
| adgrd2 | 0.021 | apodb | -0.009 |
| CU467822.1 | 0.021 | capn2b | -0.009 |
| wnt7aa | 0.021 | capn9 | -0.009 |
| XLOC-003690 | 0.021 | caspb | -0.009 |




