IGF-like family receptor 1
ZFIN 
Other cell groups


all cells |
blastomeres |
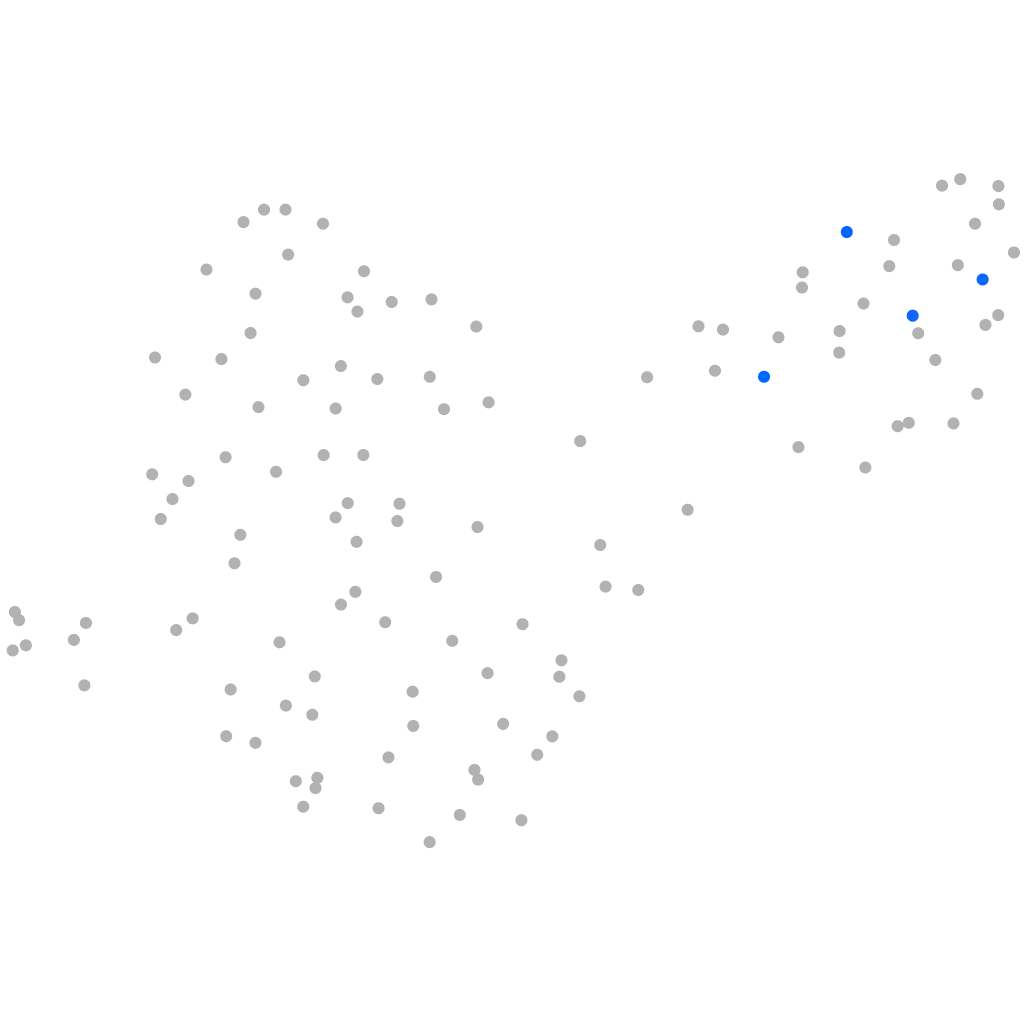
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101886464 | 0.715 | brd3a | -0.217 |
| crispld1a | 0.706 | nucks1a | -0.215 |
| pstpip1b | 0.706 | fbl | -0.212 |
| fam83ha | 0.696 | dynll2a | -0.210 |
| si:cabz01007812.1 | 0.696 | ncl | -0.209 |
| ccdc169 | 0.676 | ddx23 | -0.200 |
| ncam1b | 0.653 | u2surp | -0.200 |
| cetn2 | 0.620 | ddx18 | -0.198 |
| nfic | 0.610 | psme3 | -0.194 |
| gdpd4b | 0.589 | marcksb | -0.191 |
| LOC108179992 | 0.587 | waca | -0.190 |
| LOC108190625 | 0.580 | acin1b | -0.188 |
| rrad | 0.579 | surf2 | -0.188 |
| CT573429.2 | 0.573 | npm1a | -0.187 |
| nsmfb | 0.566 | ddit4 | -0.185 |
| zgc:158263 | 0.560 | srrm1 | -0.184 |
| stx12l | 0.550 | htatsf1 | -0.181 |
| scn1bb | 0.544 | ss18 | -0.180 |
| ANXA1 (1 of many) | 0.540 | cdc40 | -0.172 |
| si:dkey-126g1.9 | 0.536 | cnn3b | -0.171 |
| ptprz1a | 0.526 | gar1 | -0.170 |
| duox | 0.515 | dek | -0.170 |
| btbd6a | 0.514 | ybx1 | -0.168 |
| gpm6ba | 0.513 | luc7l3 | -0.166 |
| BX897691.1 | 0.510 | hmgb1a | -0.166 |
| BX936364.1 | 0.510 | tbx16 | -0.165 |
| camk1ga | 0.510 | hnrnpa1a | -0.163 |
| chia.2 | 0.510 | adssl | -0.162 |
| cntn3a.1 | 0.510 | chd4b | -0.158 |
| cntn3b | 0.510 | NC-002333.4 | -0.158 |
| CR318673.1 | 0.510 | mad2l1 | -0.156 |
| CR848746.1 | 0.510 | nob1 | -0.155 |
| cryba2a | 0.510 | epcam | -0.154 |
| crybb1 | 0.510 | ddx19 | -0.153 |
| ctgfa | 0.510 | lyar | -0.152 |