insulin-like growth factor 2 receptor
ZFIN 
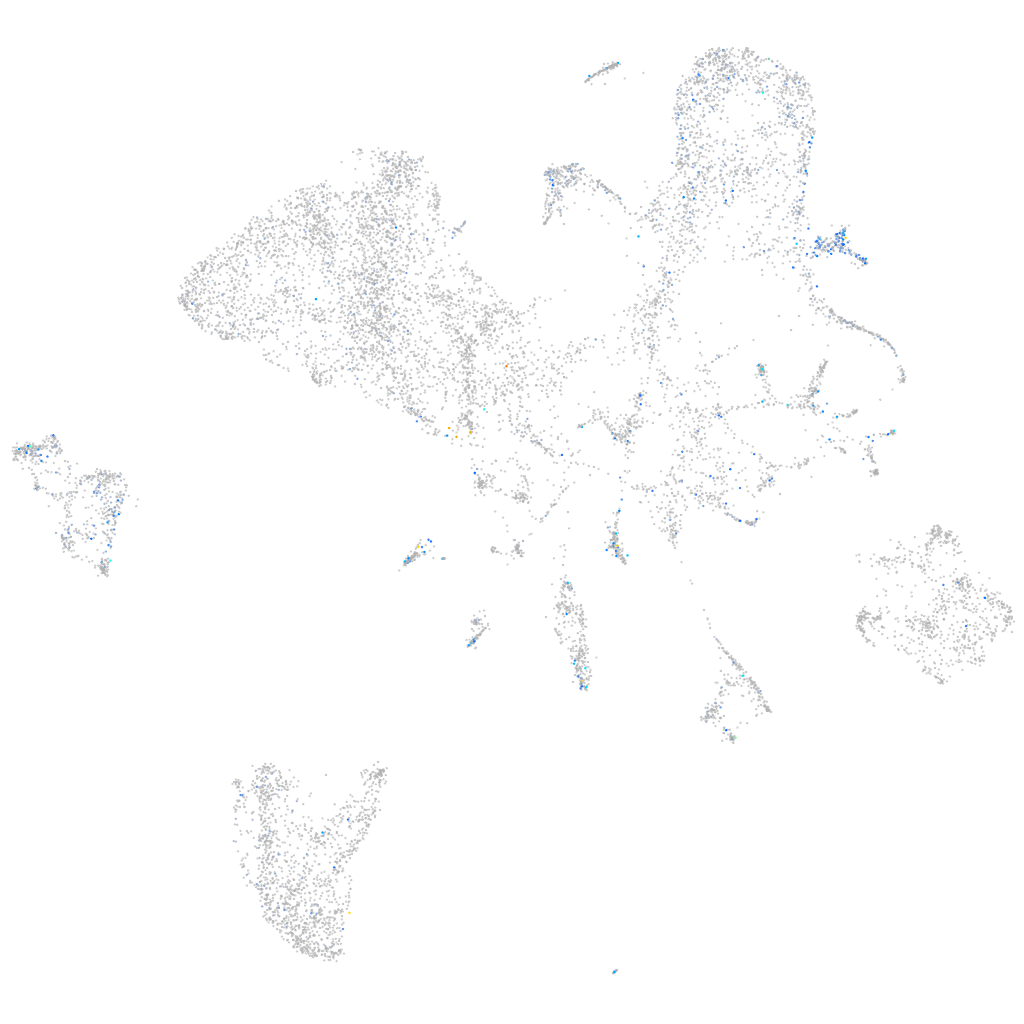
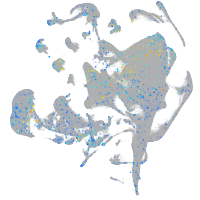
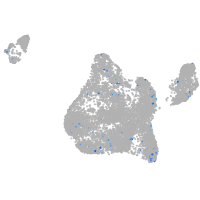
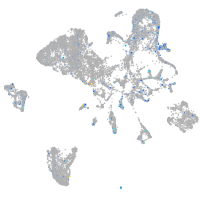
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| lrp2b | 0.188 | aqp12 | -0.098 |
| LOC100537272 | 0.187 | agxtb | -0.098 |
| ctsl.1 | 0.186 | bhmt | -0.097 |
| ctsbb | 0.182 | mat1a | -0.091 |
| cubn | 0.179 | fabp10a | -0.090 |
| naga | 0.174 | apoc1 | -0.089 |
| LOC108190565 | 0.169 | ttc36 | -0.088 |
| sptbn5 | 0.164 | rbp2b | -0.086 |
| xirp1 | 0.160 | uox | -0.086 |
| akap9 | 0.159 | grhprb | -0.085 |
| gnsb | 0.158 | kng1 | -0.085 |
| gabrz | 0.157 | ftcd | -0.083 |
| dpep1 | 0.156 | apom | -0.083 |
| myo6a | 0.155 | gc | -0.082 |
| LOC100331068 | 0.154 | ambp | -0.082 |
| fuca2 | 0.150 | cpn1 | -0.082 |
| CR381686.5 | 0.149 | hpda | -0.082 |
| amn | 0.149 | agxta | -0.081 |
| tfeb | 0.148 | serpina1 | -0.080 |
| myo3a | 0.148 | ttr | -0.080 |
| tspan34 | 0.147 | serpina1l | -0.080 |
| abcc8b | 0.146 | cfhl4 | -0.079 |
| zgc:165573 | 0.146 | pygl | -0.078 |
| epdl2 | 0.144 | zgc:123103 | -0.078 |
| dab2 | 0.142 | fgb | -0.078 |
| BX005448.1 | 0.142 | si:ch73-361h17.1 | -0.077 |
| si:ch211-266g18.9 | 0.141 | sec14l7 | -0.077 |
| si:dkey-28n18.9 | 0.140 | c9 | -0.077 |
| man2b1 | 0.140 | fga | -0.077 |
| tpte | 0.140 | tfa | -0.077 |
| lgmn | 0.139 | si:dkey-16p21.8 | -0.077 |
| CT955996.1 | 0.138 | hao1 | -0.076 |
| rbm47 | 0.138 | ces2 | -0.076 |
| scpep1 | 0.138 | serpinf2a | -0.076 |
| snx2 | 0.136 | serpinf2b | -0.076 |