"immunoglobulin superfamily, DCC subclass, member 3"
ZFIN 
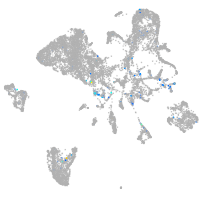
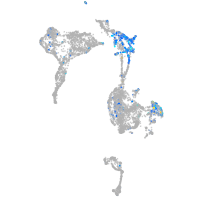
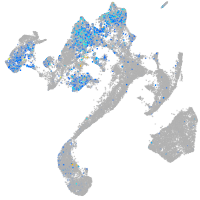
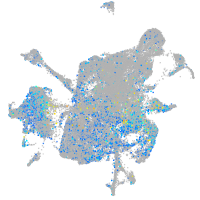
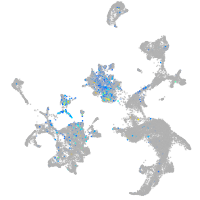
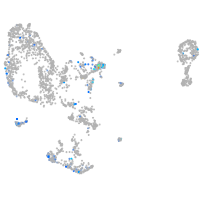
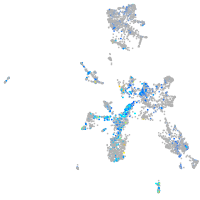
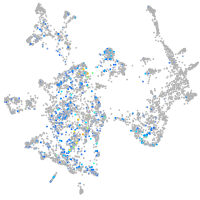
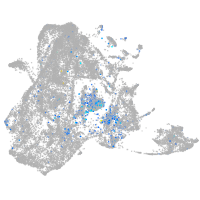
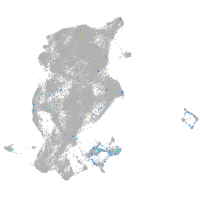
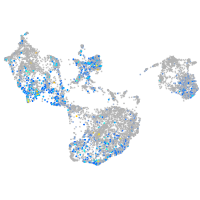
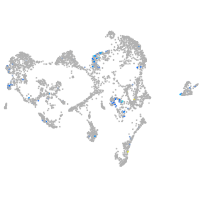
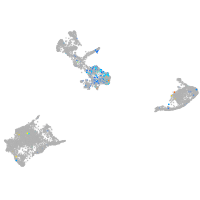
Expression by stage/cluster


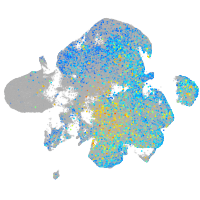
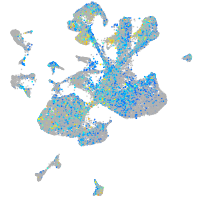
Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gpm6aa | 0.187 | cyt1 | -0.091 |
| rtn1a | 0.184 | si:ch211-195b11.3 | -0.082 |
| tmeff1b | 0.182 | krt4 | -0.070 |
| nova2 | 0.179 | cfl1l | -0.069 |
| gabbr2 | 0.176 | icn2 | -0.064 |
| gpm6ab | 0.176 | wu:fb18f06 | -0.063 |
| hmgb3a | 0.166 | cyt1l | -0.060 |
| cspg5a | 0.163 | scel | -0.060 |
| atp6v0cb | 0.163 | selenow1 | -0.058 |
| elavl3 | 0.162 | anxa1a | -0.058 |
| cdh2 | 0.159 | zgc:193505 | -0.056 |
| VSTM2A | 0.158 | aldob | -0.055 |
| si:dkeyp-75h12.5 | 0.158 | krt5 | -0.054 |
| BX005003.1 | 0.157 | pycard | -0.053 |
| tuba1c | 0.156 | s100a10b | -0.052 |
| negr1 | 0.155 | krt17 | -0.051 |
| lgi1b | 0.154 | si:dkey-16p21.8 | -0.051 |
| hmgb1b | 0.151 | zgc:162730 | -0.051 |
| vamp2 | 0.149 | glo1 | -0.050 |
| syngr3a | 0.149 | zgc:175088 | -0.049 |
| marcksl1b | 0.149 | s100v2 | -0.049 |
| elavl4 | 0.147 | mgst1.2 | -0.048 |
| CABZ01029822.1 | 0.147 | krtt1c19e | -0.047 |
| marcksl1a | 0.147 | eef1da | -0.046 |
| tubb5 | 0.147 | fbp1a | -0.046 |
| myt1la | 0.147 | anxa1c | -0.045 |
| lhx1a | 0.147 | rpl35a | -0.045 |
| rtn1b | 0.145 | selenow2b | -0.045 |
| agrn | 0.143 | si:ch73-52f15.5 | -0.044 |
| cacng4b | 0.143 | cst14b.1 | -0.043 |
| plxna1a | 0.143 | si:ch211-157c3.4 | -0.043 |
| mdkb | 0.142 | icn | -0.043 |
| brinp3a.1 | 0.142 | tcima | -0.042 |
| sprn | 0.141 | agr1 | -0.042 |
| stmn1b | 0.141 | si:ch211-105c13.3 | -0.041 |