immunoglobulin (CD79A) binding protein 1
ZFIN 
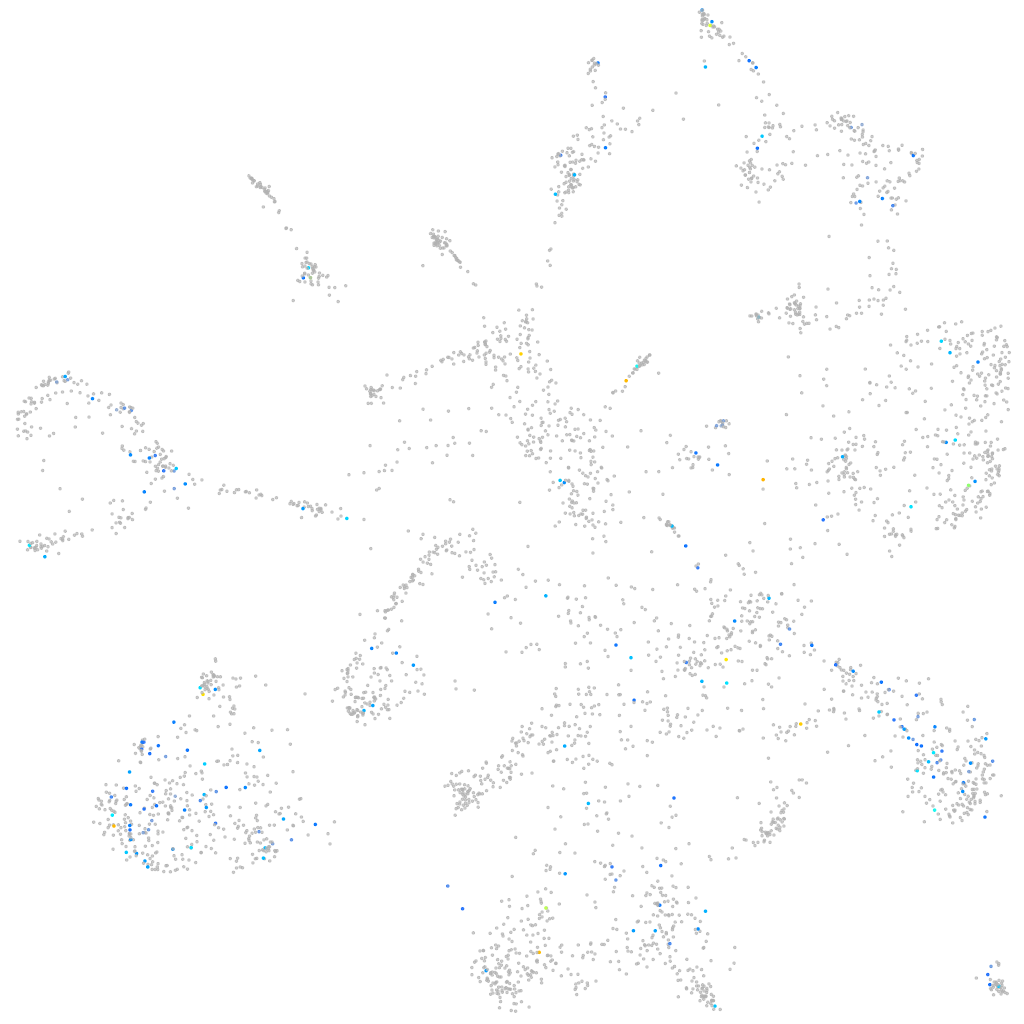
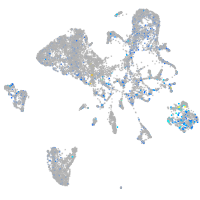
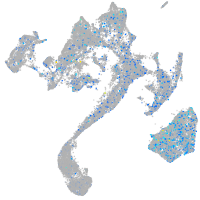
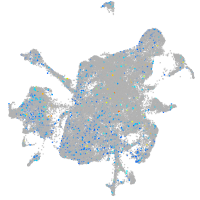
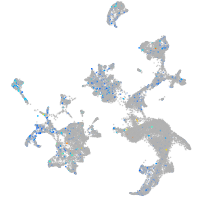
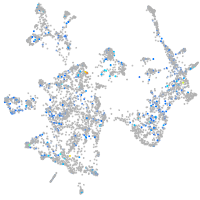
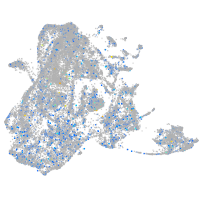
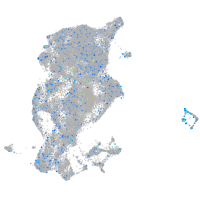
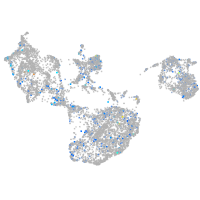
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| sptssb | 0.173 | col1a2 | -0.050 |
| LOC110440146 | 0.161 | col5a1 | -0.043 |
| LOC103911479 | 0.144 | pvalb2 | -0.043 |
| BX569796.1 | 0.134 | rbms2a | -0.042 |
| fam124b | 0.130 | rplp2 | -0.041 |
| usp21 | 0.128 | rbp4 | -0.040 |
| LOC100334495 | 0.128 | mmp15a | -0.038 |
| slc25a23b | 0.128 | si:ch211-250c4.4 | -0.037 |
| kcnq3 | 0.126 | rhag | -0.037 |
| helq | 0.126 | krt8 | -0.037 |
| cldn34a | 0.124 | actc1b | -0.036 |
| mmp16b | 0.122 | pcolce2b | -0.036 |
| prickle2a | 0.121 | vim | -0.036 |
| nt5c1aa | 0.120 | nid1a | -0.035 |
| CU459186.3 | 0.119 | tspan4a | -0.035 |
| XLOC-018221 | 0.119 | ccdc80 | -0.035 |
| znf1108 | 0.116 | edem1 | -0.034 |
| LOC110439309 | 0.116 | slit3 | -0.033 |
| neto1l | 0.115 | fn1a | -0.033 |
| LOC101882298 | 0.115 | si:dkey-28a3.2 | -0.033 |
| XLOC-031067 | 0.114 | pvalb1 | -0.033 |
| CU855688.2 | 0.113 | wdr33 | -0.032 |
| cabp7b | 0.113 | ckmb | -0.032 |
| LOC562098 | 0.112 | LOC101883507 | -0.031 |
| mep1a.2 | 0.112 | ahnak | -0.031 |
| CABZ01086139.1 | 0.112 | myl13 | -0.031 |
| gria4a | 0.112 | dhrs1 | -0.031 |
| LOC101887100 | 0.111 | ifitm1 | -0.030 |
| lrrc73 | 0.111 | myhz1.1 | -0.030 |
| slc7a7 | 0.109 | mir199-3 | -0.030 |
| wdr92 | 0.109 | col18a1a | -0.030 |
| nmnat2 | 0.107 | dcun1d4 | -0.030 |
| zgc:152658 | 0.106 | ppp2r3a | -0.030 |
| CABZ01066312.1 | 0.106 | sparc | -0.030 |
| CT573316.1 | 0.105 | pik3cb | -0.030 |