intraflagellar transport 22 homolog (Chlamydomonas)
ZFIN 
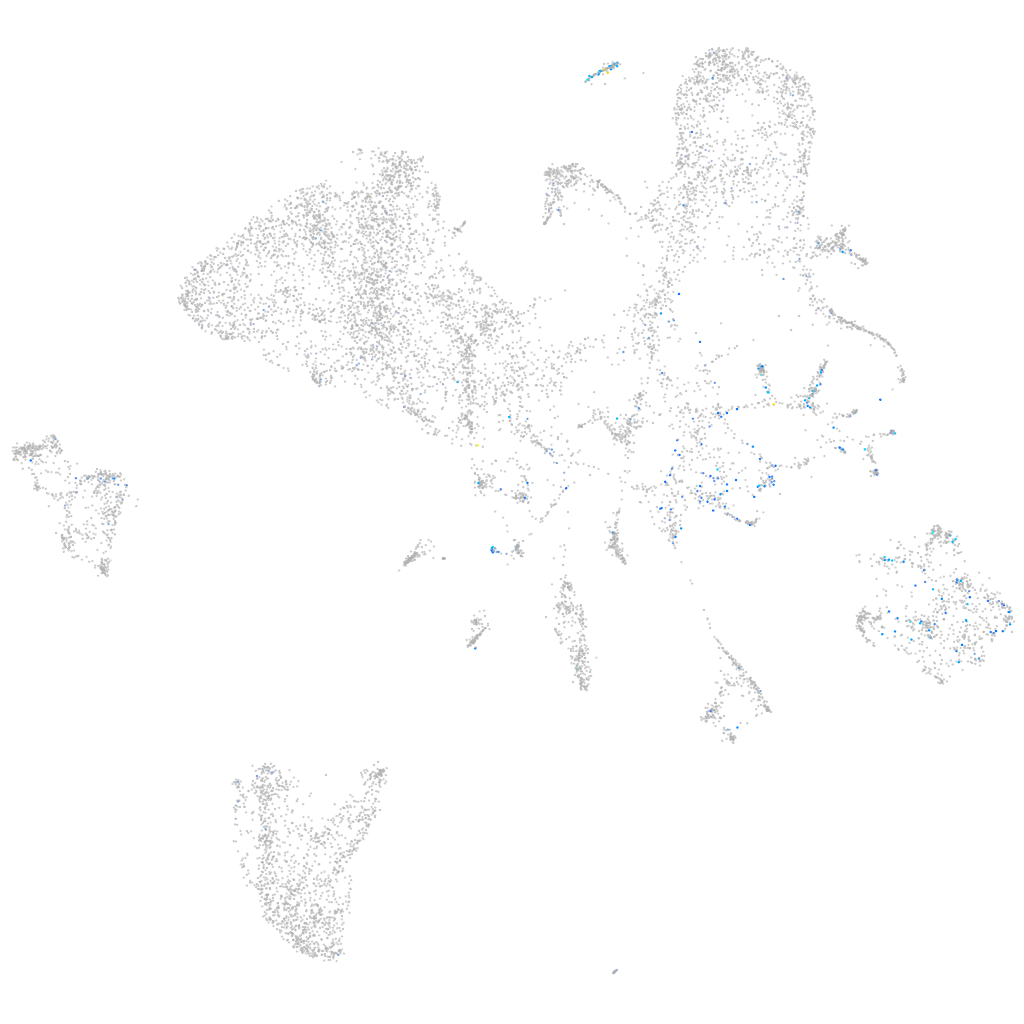
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-42p14.3 | 0.197 | gapdh | -0.130 |
| si:dkey-11c5.11 | 0.191 | gamt | -0.129 |
| spata18 | 0.191 | ahcy | -0.129 |
| insm1b | 0.176 | gatm | -0.120 |
| scg3 | 0.176 | nupr1b | -0.108 |
| egr4 | 0.174 | bhmt | -0.106 |
| capsla | 0.173 | fbp1b | -0.104 |
| penka | 0.173 | dap | -0.103 |
| zgc:101731 | 0.168 | mat1a | -0.103 |
| neurod1 | 0.160 | eno3 | -0.102 |
| zgc:195356 | 0.158 | aldob | -0.102 |
| adcyap1a | 0.158 | aldh6a1 | -0.102 |
| pih1d3 | 0.157 | cx32.3 | -0.101 |
| CR855277.2 | 0.157 | apoa4b.1 | -0.101 |
| lysmd2 | 0.155 | apoa1b | -0.101 |
| fev | 0.152 | afp4 | -0.097 |
| erich2 | 0.149 | apoc2 | -0.096 |
| hmgn6 | 0.149 | scp2a | -0.096 |
| efcab2 | 0.148 | glud1b | -0.095 |
| meig1 | 0.147 | suclg1 | -0.094 |
| cacna2d4a | 0.146 | fabp3 | -0.094 |
| si:ch211-222l21.1 | 0.145 | gnmt | -0.094 |
| si:ch211-147d7.5 | 0.143 | agxtb | -0.094 |
| si:ch73-160i9.2 | 0.140 | gstt1a | -0.093 |
| hnrnpaba | 0.140 | aqp12 | -0.093 |
| ap3s2 | 0.139 | abat | -0.091 |
| lmx1ba | 0.139 | mgst1.2 | -0.090 |
| capslb | 0.139 | eef1da | -0.090 |
| ret | 0.137 | apoa2 | -0.089 |
| pacrg | 0.137 | aldh7a1 | -0.089 |
| tspan7 | 0.136 | pgm1 | -0.089 |
| rgs16 | 0.136 | pnp4b | -0.089 |
| h3f3d | 0.136 | mdh1aa | -0.087 |
| si:ch73-222h13.1 | 0.133 | apobb.1 | -0.087 |
| nkx2.2a | 0.132 | suclg2 | -0.086 |