isocitrate dehydrogenase (NAD(+)) 3 catalytic subunit alpha
ZFIN 
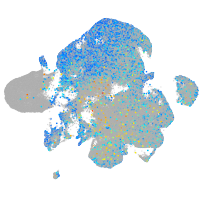
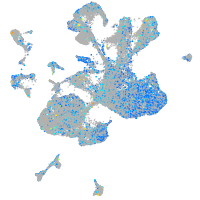
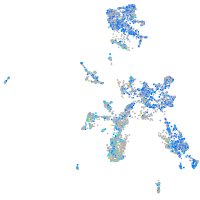
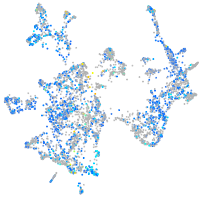
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tnrc6a | 0.631 | NC-002333.4 | -0.552 |
| npm2b | 0.618 | ncl | -0.526 |
| calm3a | 0.618 | fbl | -0.481 |
| hnrnpa0l | 0.611 | hnrnpa1b | -0.474 |
| cct2 | 0.600 | marcksl1b | -0.447 |
| rps10 | 0.590 | tbx16 | -0.437 |
| rpl37 | 0.589 | ppig | -0.437 |
| rps17 | 0.584 | cdk11b | -0.426 |
| mrpl33 | 0.577 | hnrnpub | -0.420 |
| zgc:114188 | 0.576 | acin1b | -0.417 |
| tdrkh | 0.574 | akap12b | -0.405 |
| FQ976913.1 | 0.571 | hnrnpa1a | -0.404 |
| srsf7b | 0.570 | srrt | -0.399 |
| sipa1l2 | 0.563 | nop58 | -0.398 |
| dap1b | 0.561 | snrnp70 | -0.395 |
| enosf1 | 0.561 | top1l | -0.392 |
| cib1 | 0.558 | npm1a | -0.389 |
| zgc:171566 | 0.556 | marcksb | -0.387 |
| mif | 0.555 | slu7 | -0.384 |
| ISCU (1 of many) | 0.554 | cactin | -0.383 |
| pon3.2 | 0.553 | ISCU | -0.381 |
| txndc15 | 0.553 | tpx2 | -0.379 |
| golgb1 | 0.547 | h1m | -0.377 |
| cox15 | 0.544 | cd2bp2 | -0.377 |
| hyi | 0.536 | brd3a | -0.371 |
| tmem243b | 0.533 | anp32a | -0.371 |
| btf3 | 0.532 | elavl1a | -0.366 |
| zgc:56493 | 0.532 | banf1 | -0.363 |
| rpl19 | 0.530 | stm | -0.359 |
| exosc3 | 0.530 | srrm1 | -0.354 |
| gtf2h4 | 0.529 | nasp | -0.353 |
| dusp22b | 0.529 | rps18 | -0.352 |
| rnf175 | 0.526 | hnrnpaba | -0.352 |
| prdx2 | 0.519 | baz1b | -0.349 |
| marcksl1a | 0.518 | gra | -0.339 |