heat shock protein b8
ZFIN 
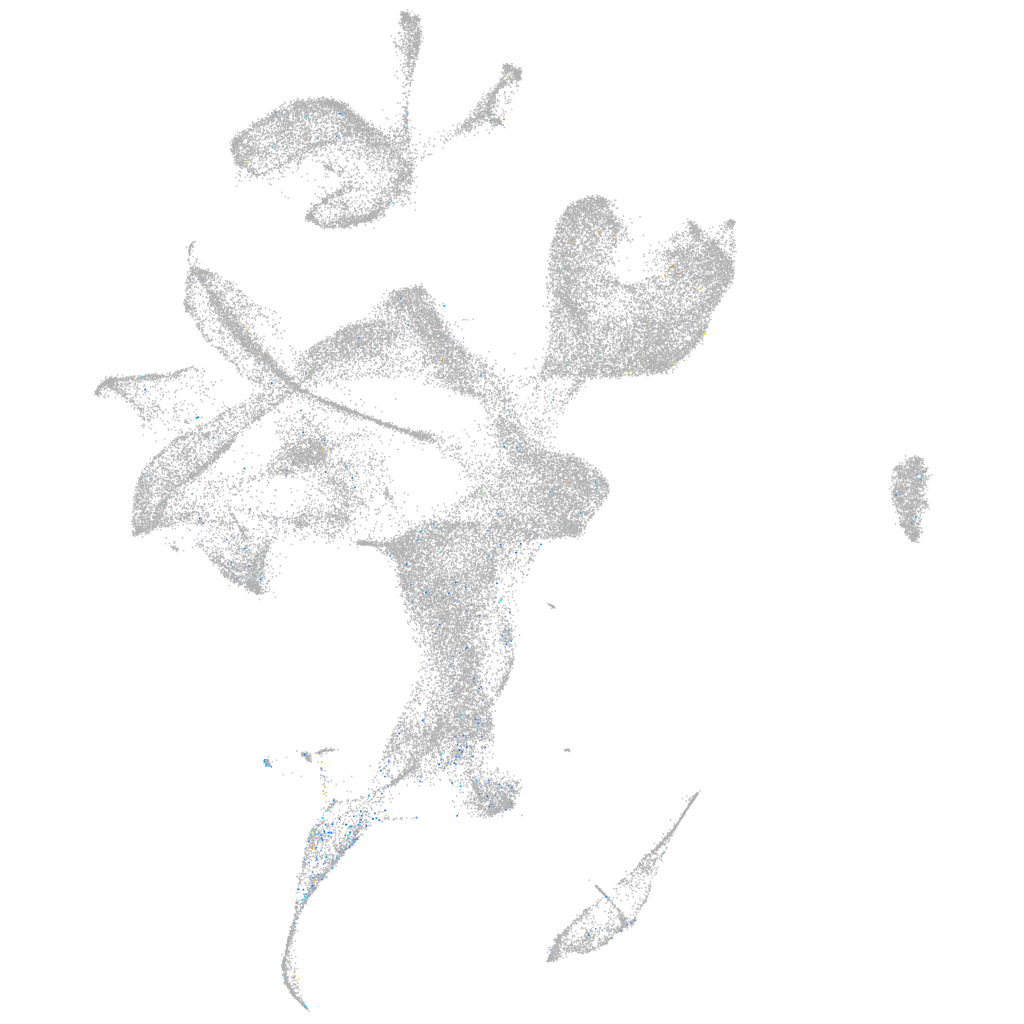
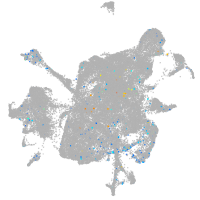
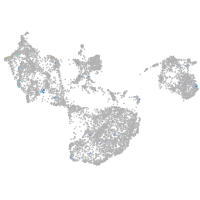
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tfec | 0.134 | ptmab | -0.059 |
| rab32a | 0.123 | gpm6aa | -0.042 |
| ednrab | 0.122 | ptmaa | -0.040 |
| tspan36 | 0.120 | si:ch73-1a9.3 | -0.040 |
| qdpra | 0.118 | tuba1c | -0.040 |
| LOC101885388 | 0.114 | hmgb1a | -0.038 |
| slc45a2 | 0.111 | hmgn6 | -0.037 |
| trpm1b | 0.110 | ckbb | -0.037 |
| rab38 | 0.109 | cadm3 | -0.034 |
| cax1 | 0.109 | gapdhs | -0.034 |
| gpr143 | 0.107 | fabp7a | -0.031 |
| zgc:110591 | 0.101 | atp6v0cb | -0.031 |
| pmela | 0.100 | snap25b | -0.031 |
| dct | 0.099 | gpm6ab | -0.030 |
| pttg1ipb | 0.095 | si:ch211-137a8.4 | -0.029 |
| amdhd2 | 0.093 | nova2 | -0.028 |
| pcbd1 | 0.091 | sncb | -0.027 |
| agtrap | 0.091 | atp1a3a | -0.027 |
| tyrp1b | 0.090 | foxg1b | -0.027 |
| bcl3 | 0.090 | rnasekb | -0.026 |
| slc15a2 | 0.090 | rtn1a | -0.026 |
| mitfa | 0.089 | crx | -0.025 |
| msx1b | 0.089 | atpv0e2 | -0.025 |
| rnaseka | 0.089 | mid1ip1b | -0.025 |
| crestin | 0.089 | anp32e | -0.025 |
| gstp1 | 0.089 | btg1 | -0.025 |
| FP085398.1 | 0.088 | cspg5a | -0.024 |
| tpm4a | 0.086 | ppiab | -0.024 |
| tyrp1a | 0.085 | syt5b | -0.024 |
| tmem88b | 0.084 | tuba1a | -0.024 |
| XLOC-004932 | 0.084 | stmn1b | -0.024 |
| sox10 | 0.083 | vamp2 | -0.024 |
| sytl2b | 0.083 | mpp6b | -0.024 |
| tyr | 0.082 | rorb | -0.024 |
| vaspa | 0.082 | sypb | -0.024 |