"heat shock protein 90, alpha (cytosolic), class A member 1, tandem duplicate 2"
ZFIN 
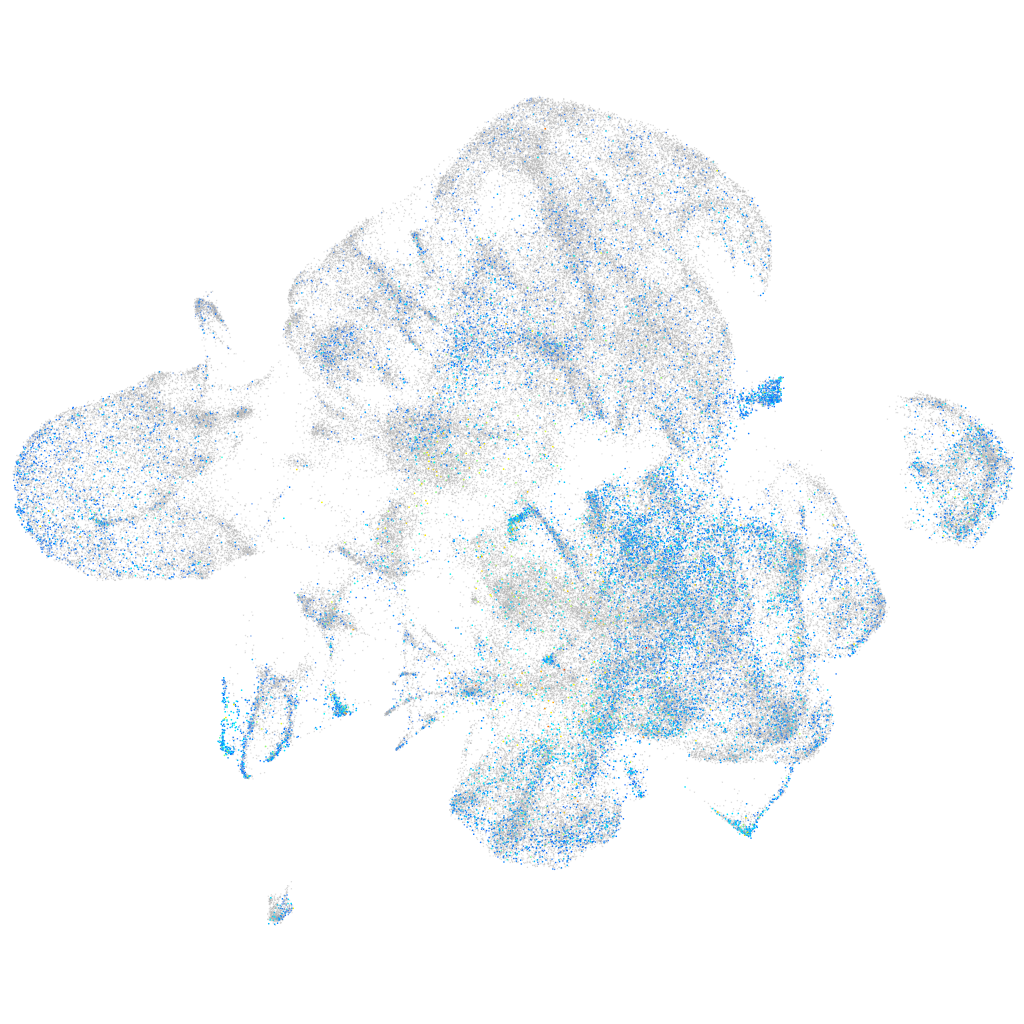
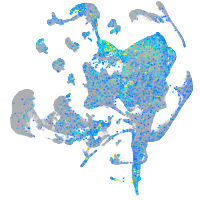
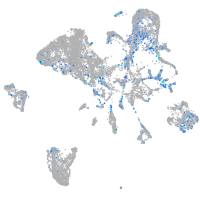
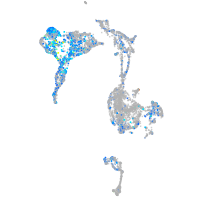
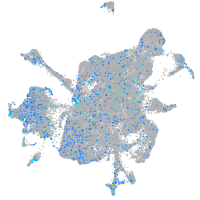
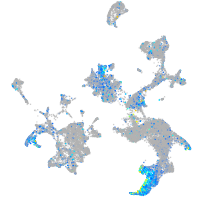
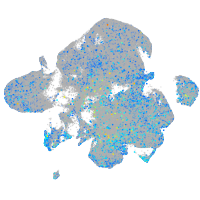
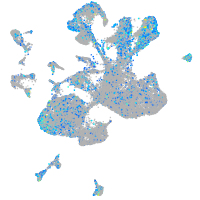
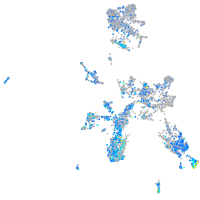
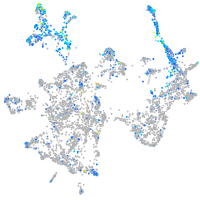
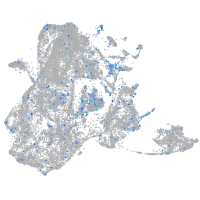
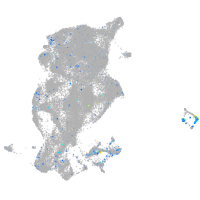
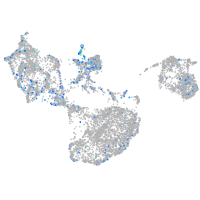
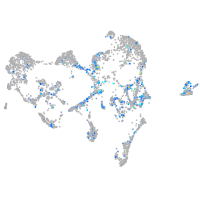
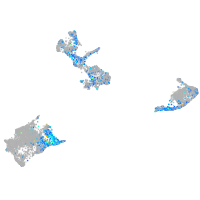
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| gapdhs | 0.151 | XLOC-003690 | -0.112 |
| ckbb | 0.148 | id1 | -0.108 |
| atp6v1e1b | 0.141 | mdka | -0.105 |
| gng3 | 0.137 | XLOC-003689 | -0.104 |
| mllt11 | 0.136 | sox3 | -0.099 |
| vamp2 | 0.136 | cldn5a | -0.098 |
| stxbp1a | 0.134 | rplp1 | -0.097 |
| calm2a | 0.133 | si:dkey-151g10.6 | -0.095 |
| rnasekb | 0.130 | hmgb2a | -0.094 |
| zgc:65894 | 0.130 | notch3 | -0.091 |
| atp6v1g1 | 0.128 | gfap | -0.089 |
| atp6v0cb | 0.128 | her2 | -0.088 |
| ywhag2 | 0.128 | XLOC-003692 | -0.087 |
| sncb | 0.128 | sox19a | -0.084 |
| cnrip1a | 0.127 | rps20 | -0.083 |
| si:dkey-280e21.3 | 0.125 | sdc4 | -0.083 |
| stx1b | 0.123 | msi1 | -0.083 |
| tuba1c | 0.123 | si:dkey-42i9.4 | -0.082 |
| gnao1a | 0.123 | COX7A2 (1 of many) | -0.082 |
| gdi1 | 0.122 | sb:cb81 | -0.082 |
| vat1 | 0.122 | pcna | -0.082 |
| calm1a | 0.121 | her15.1 | -0.082 |
| gng2 | 0.121 | lfng | -0.081 |
| atp5mc1 | 0.119 | zfp36l1a | -0.080 |
| gnb1a | 0.118 | si:ch211-212k18.5 | -0.080 |
| stmn1b | 0.118 | abhd6a | -0.080 |
| elavl4 | 0.117 | rps12 | -0.079 |
| csdc2a | 0.117 | tuba8l | -0.079 |
| stmn2a | 0.116 | her9 | -0.078 |
| tpi1b | 0.115 | selenoh | -0.077 |
| dynll1 | 0.115 | nrarpb | -0.076 |
| rtn1b | 0.115 | si:ch73-21g5.7 | -0.074 |
| si:dkeyp-75h12.5 | 0.114 | her4.1 | -0.074 |
| zgc:153426 | 0.113 | rplp2l | -0.074 |
| calr | 0.113 | btg2 | -0.073 |