heat shock factor binding protein 1a
ZFIN 
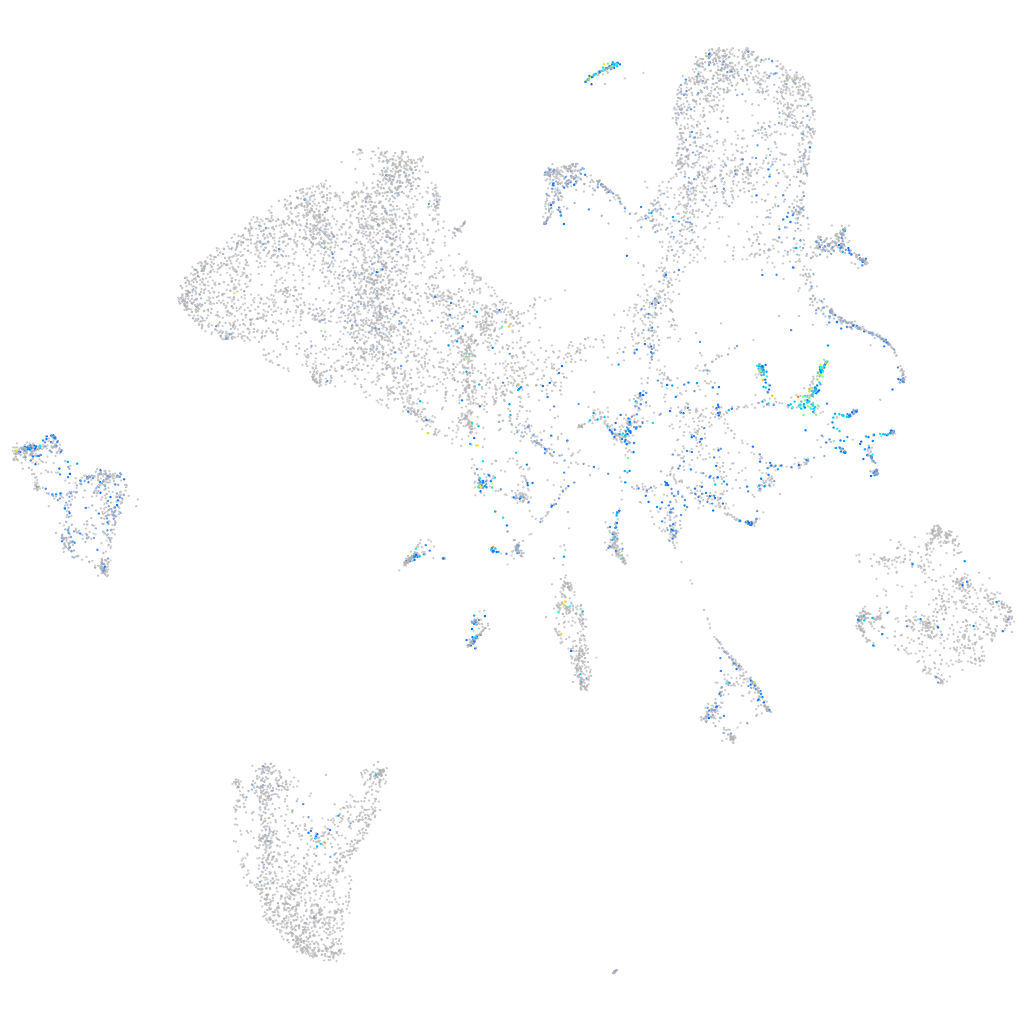
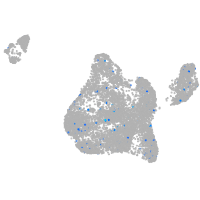
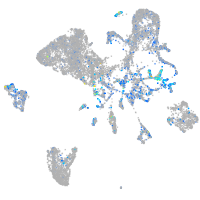
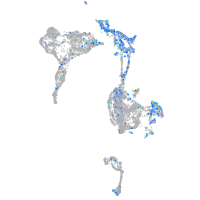
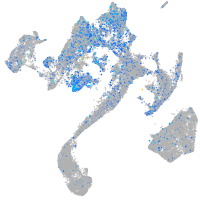
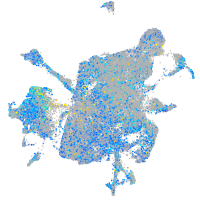
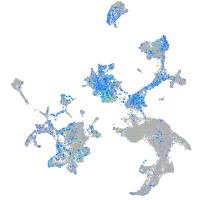
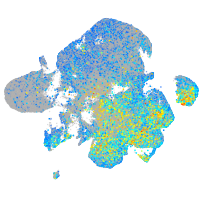
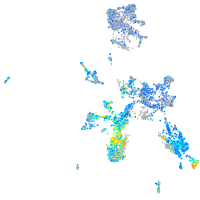
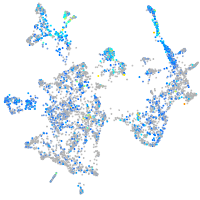
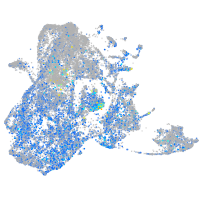
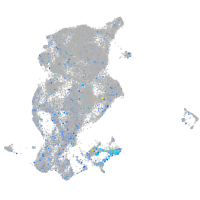
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| scg3 | 0.425 | gamt | -0.243 |
| egr4 | 0.371 | gatm | -0.235 |
| arxa | 0.367 | gapdh | -0.231 |
| neurod1 | 0.362 | ahcy | -0.213 |
| fev | 0.361 | mat1a | -0.212 |
| scgn | 0.353 | bhmt | -0.212 |
| pcsk1nl | 0.350 | fbp1b | -0.202 |
| mir7a-1 | 0.349 | agxtb | -0.192 |
| calm1b | 0.347 | nupr1b | -0.191 |
| capsla | 0.346 | aqp12 | -0.190 |
| insm1b | 0.343 | apoa1b | -0.189 |
| c2cd4a | 0.343 | cx32.3 | -0.187 |
| gapdhs | 0.327 | apoa4b.1 | -0.186 |
| syt1a | 0.322 | aldob | -0.184 |
| rnasekb | 0.321 | aldh6a1 | -0.181 |
| id4 | 0.318 | pnp4b | -0.180 |
| pax6b | 0.317 | apoc1 | -0.179 |
| vat1 | 0.317 | afp4 | -0.179 |
| hepacam2 | 0.316 | si:dkey-16p21.8 | -0.178 |
| si:ch1073-429i10.3.1 | 0.314 | apoc2 | -0.177 |
| tmsb | 0.310 | glud1b | -0.177 |
| anxa4 | 0.305 | abat | -0.173 |
| rasd4 | 0.302 | scp2a | -0.173 |
| si:dkey-153k10.9 | 0.299 | ttc36 | -0.171 |
| elovl1a | 0.298 | hdlbpa | -0.170 |
| insm1a | 0.298 | agxta | -0.169 |
| ptmaa | 0.292 | grhprb | -0.167 |
| zgc:101731 | 0.292 | fabp10a | -0.166 |
| calm1a | 0.291 | apom | -0.165 |
| h3f3c | 0.290 | g6pca.2 | -0.165 |
| vamp2 | 0.290 | ftcd | -0.165 |
| mir375-2 | 0.288 | tfa | -0.165 |
| dll4 | 0.286 | aldh7a1 | -0.165 |
| si:ch73-160i9.2 | 0.286 | rbp2b | -0.164 |
| ap3s2 | 0.281 | gstt1a | -0.164 |