heparan sulfate (glucosamine) 3-O-sulfotransferase 1-like 2
ZFIN 
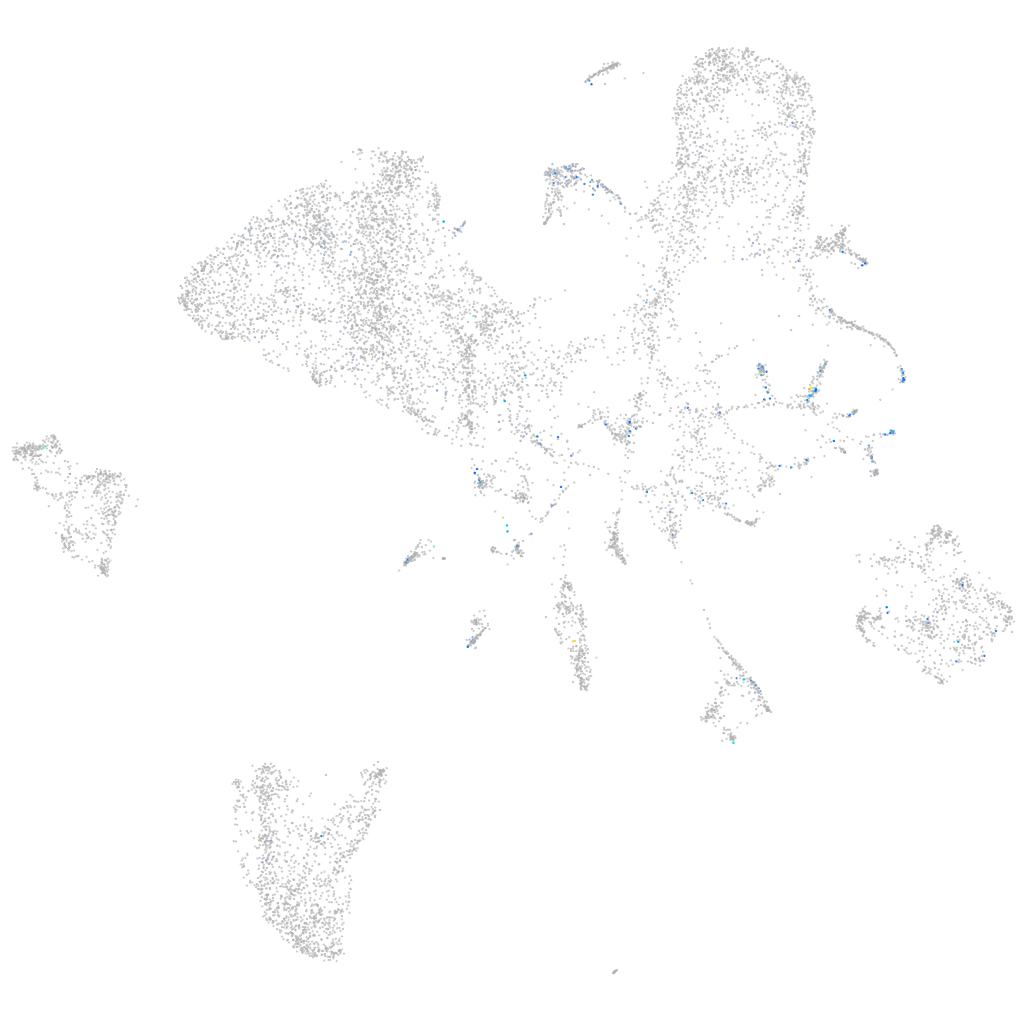
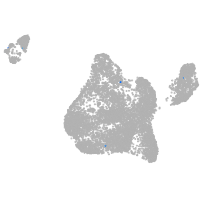
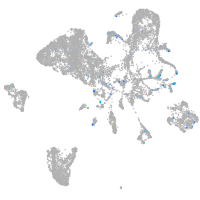
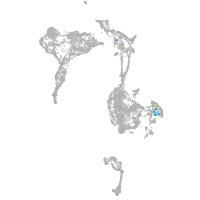
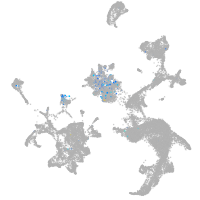
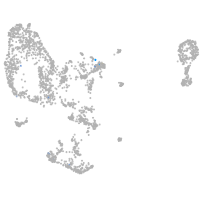
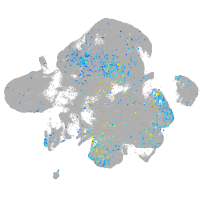
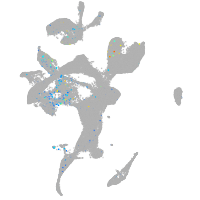
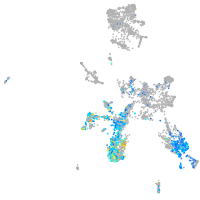
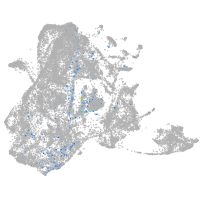
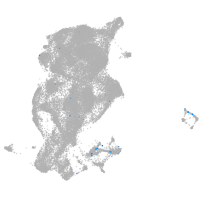
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CABZ01058333.1 | 0.189 | ahcy | -0.090 |
| XLOC-038255 | 0.166 | gamt | -0.088 |
| LOC110439360 | 0.158 | gapdh | -0.084 |
| BX545856.1 | 0.155 | gatm | -0.077 |
| scg3 | 0.149 | fbp1b | -0.076 |
| vat1 | 0.146 | mat1a | -0.076 |
| rem2 | 0.144 | hdlbpa | -0.075 |
| il26 | 0.143 | dap | -0.072 |
| gfra3 | 0.136 | apoa4b.1 | -0.072 |
| rasd4 | 0.135 | glud1b | -0.070 |
| syt1a | 0.132 | sult2st2 | -0.070 |
| pbx1a | 0.130 | apoc2 | -0.070 |
| camk2n1a | 0.129 | gstt1a | -0.070 |
| calm1b | 0.129 | rgn | -0.067 |
| si:ch211-153b23.3 | 0.127 | suclg1 | -0.067 |
| anxa4 | 0.127 | aqp12 | -0.067 |
| XLOC-002962 | 0.125 | gstr | -0.067 |
| elovl1a | 0.123 | pnp4b | -0.066 |
| neurod1 | 0.123 | bhmt | -0.066 |
| taar20f | 0.123 | scp2a | -0.065 |
| pcsk1nl | 0.122 | agxtb | -0.064 |
| TCIM (1 of many) | 0.122 | mgst1.2 | -0.064 |
| scgn | 0.121 | rdh1 | -0.064 |
| adora2aa | 0.121 | cx32.3 | -0.064 |
| kcnk9 | 0.121 | nupr1b | -0.063 |
| muc5e | 0.121 | dhrs9 | -0.062 |
| FP016005.1 | 0.120 | eif4ebp3 | -0.061 |
| fev | 0.120 | apobb.1 | -0.061 |
| BX247868.1 | 0.120 | aldh7a1 | -0.060 |
| pitpnaa | 0.120 | afp4 | -0.060 |
| rprmb | 0.119 | aldh6a1 | -0.059 |
| tspan7b | 0.119 | nipsnap3a | -0.059 |
| poln | 0.119 | si:dkey-16p21.8 | -0.059 |
| gapdhs | 0.119 | tdo2a | -0.059 |
| capns1b | 0.118 | apoa1b | -0.058 |