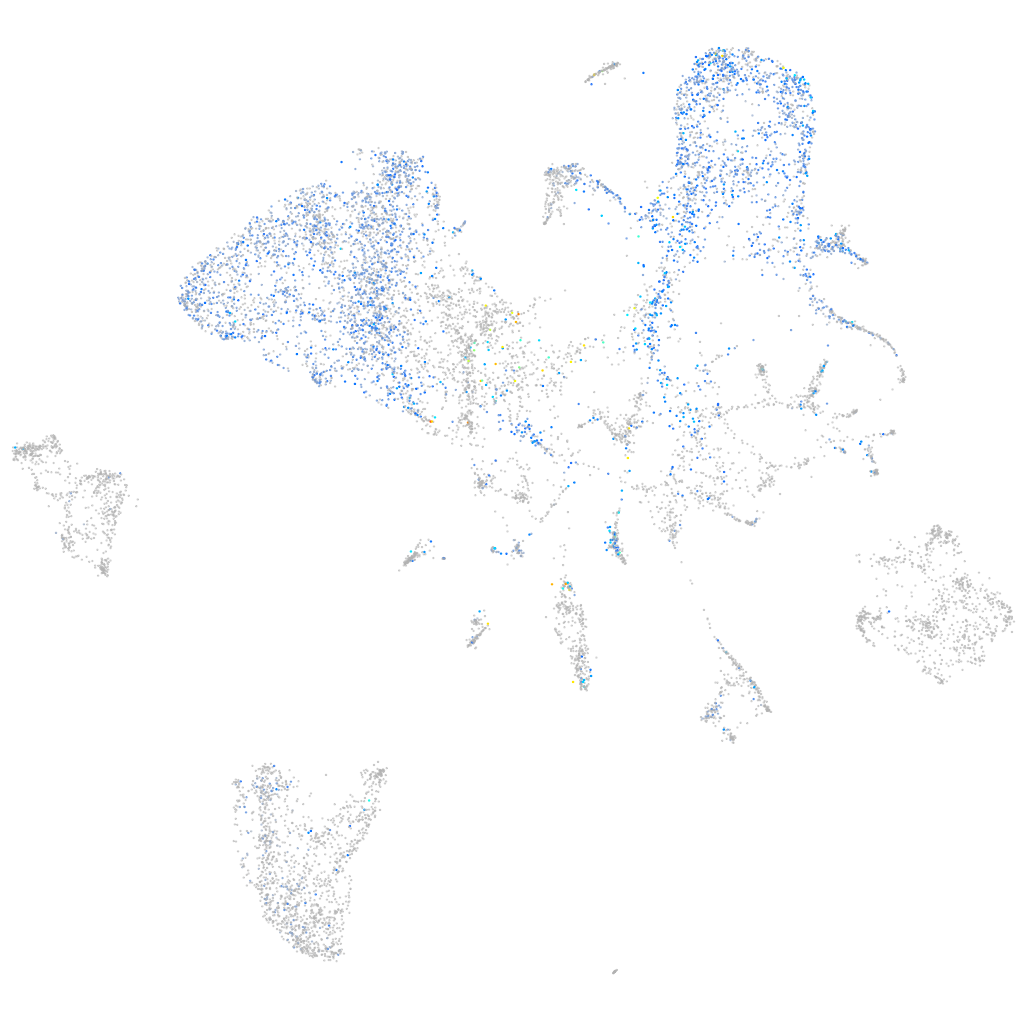
"hepatocyte nuclear factor 4, alpha"
ZFIN 
Other cell groups


all cells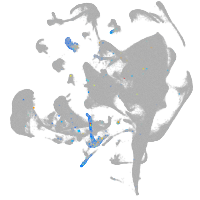 |
blastomeres |
PGCs |
endoderm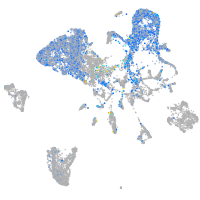 |
axial mesoderm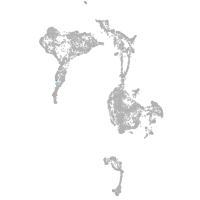 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ugt1a7 | 0.329 | si:ch211-195b11.3 | -0.185 |
| sult2st2 | 0.310 | marcksl1b | -0.171 |
| mttp | 0.308 | rtn1a | -0.171 |
| gstr | 0.305 | ctrl | -0.160 |
| abcc2 | 0.302 | prss59.2 | -0.156 |
| eno3 | 0.302 | hspb1 | -0.152 |
| creb3l3a | 0.298 | slc38a5b | -0.152 |
| gstt1a | 0.297 | ela2l | -0.152 |
| ndrg1a | 0.296 | ctrb1 | -0.152 |
| prdx2 | 0.295 | zgc:112160 | -0.152 |
| srd5a2a | 0.291 | akap12b | -0.151 |
| clic5b | 0.290 | prss59.1 | -0.151 |
| cyp2y3 | 0.286 | CELA1 (1 of many) | -0.150 |
| CU682777.2 | 0.286 | cpa5 | -0.150 |
| glud1b | 0.284 | ela3l | -0.150 |
| cox6b1 | 0.283 | cpb1 | -0.150 |
| sdr16c5b | 0.283 | sycn.2 | -0.150 |
| aco1 | 0.283 | prss1 | -0.149 |
| abcg2a | 0.282 | ela2 | -0.149 |
| ezra | 0.281 | fep15 | -0.148 |
| gsta.1 | 0.280 | zgc:136461 | -0.148 |
| fdx1 | 0.279 | apoda.2 | -0.147 |
| si:ch211-201h21.5 | 0.275 | erp27 | -0.146 |
| si:dkeyp-73b11.8 | 0.274 | pdia2 | -0.145 |
| rmdn1 | 0.272 | cel.1 | -0.145 |
| rdh1 | 0.271 | c6ast4 | -0.144 |
| cyp8b1 | 0.270 | marcksb | -0.143 |
| slco1d1 | 0.270 | cpa4 | -0.143 |
| mdh1aa | 0.270 | si:ch211-240l19.5 | -0.143 |
| gcshb | 0.268 | cx43.4 | -0.143 |
| atp5f1b | 0.268 | cel.2 | -0.142 |
| dgat2 | 0.266 | glulb | -0.142 |
| suclg1 | 0.264 | si:ch211-152c2.3 | -0.142 |
| acsf2 | 0.263 | zgc:92137 | -0.140 |
| slc37a4a | 0.261 | amy2a | -0.137 |