helicase-like transcription factor
ZFIN 
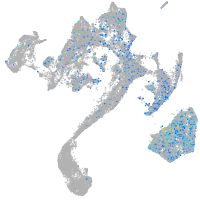
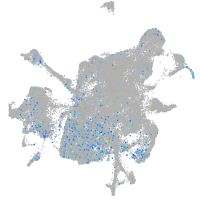
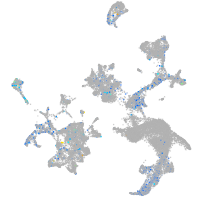
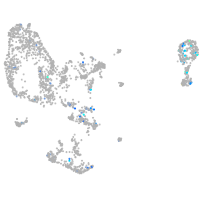
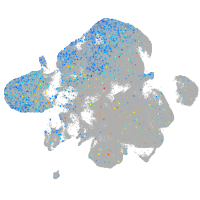
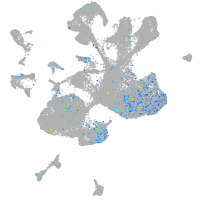
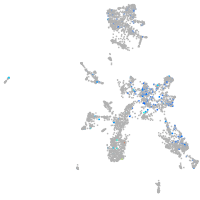
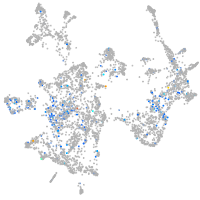
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:114120 | 0.229 | txn | -0.094 |
| BX629344.1 | 0.217 | fthl27 | -0.080 |
| XLOC-030902 | 0.202 | anxa5b | -0.080 |
| LOC100006752 | 0.183 | wu:fb18f06 | -0.072 |
| rpa3 | 0.179 | cldnh | -0.072 |
| si:ch73-252i11.1 | 0.174 | zgc:165573 | -0.067 |
| zgc:110540 | 0.170 | gapdhs | -0.067 |
| rpa2 | 0.167 | bik | -0.066 |
| dut | 0.166 | selenow1 | -0.066 |
| CABZ01005379.1 | 0.165 | calm1a | -0.066 |
| lig1 | 0.164 | ap1s3b | -0.065 |
| or126-5 | 0.162 | tmem59 | -0.063 |
| pcna | 0.160 | atp6v1e1b | -0.063 |
| chtf8 | 0.159 | h1f0 | -0.063 |
| fen1 | 0.159 | kif5bb | -0.061 |
| si:ch211-156b7.4 | 0.156 | tnks1bp1 | -0.061 |
| rrm1 | 0.155 | itm2ba | -0.061 |
| stmn1a | 0.155 | b2ml | -0.061 |
| si:dkey-6i22.5 | 0.151 | gstp1 | -0.060 |
| asf1ba | 0.151 | fxyd1 | -0.059 |
| rrm2 | 0.150 | serinc1 | -0.059 |
| nasp | 0.150 | gng3 | -0.057 |
| dnmt1 | 0.150 | rnasekb | -0.057 |
| dhfr | 0.149 | si:ch1073-443f11.2 | -0.056 |
| rfc2 | 0.149 | tmbim4 | -0.056 |
| mcm7 | 0.148 | calm1b | -0.056 |
| cks1b | 0.148 | arfgap3 | -0.056 |
| mibp | 0.148 | klf7a | -0.056 |
| chpfb | 0.148 | gbgt1l4 | -0.056 |
| chaf1a | 0.148 | CR383676.1 | -0.055 |
| cenps | 0.147 | eml2 | -0.055 |
| esco2 | 0.146 | canx | -0.055 |
| shmt1 | 0.146 | COX3 | -0.055 |
| ccna2 | 0.145 | atp6v0cb | -0.054 |
| LOC100330864 | 0.144 | bada | -0.054 |