histone cluster 1 H2A family member 9
ZFIN 
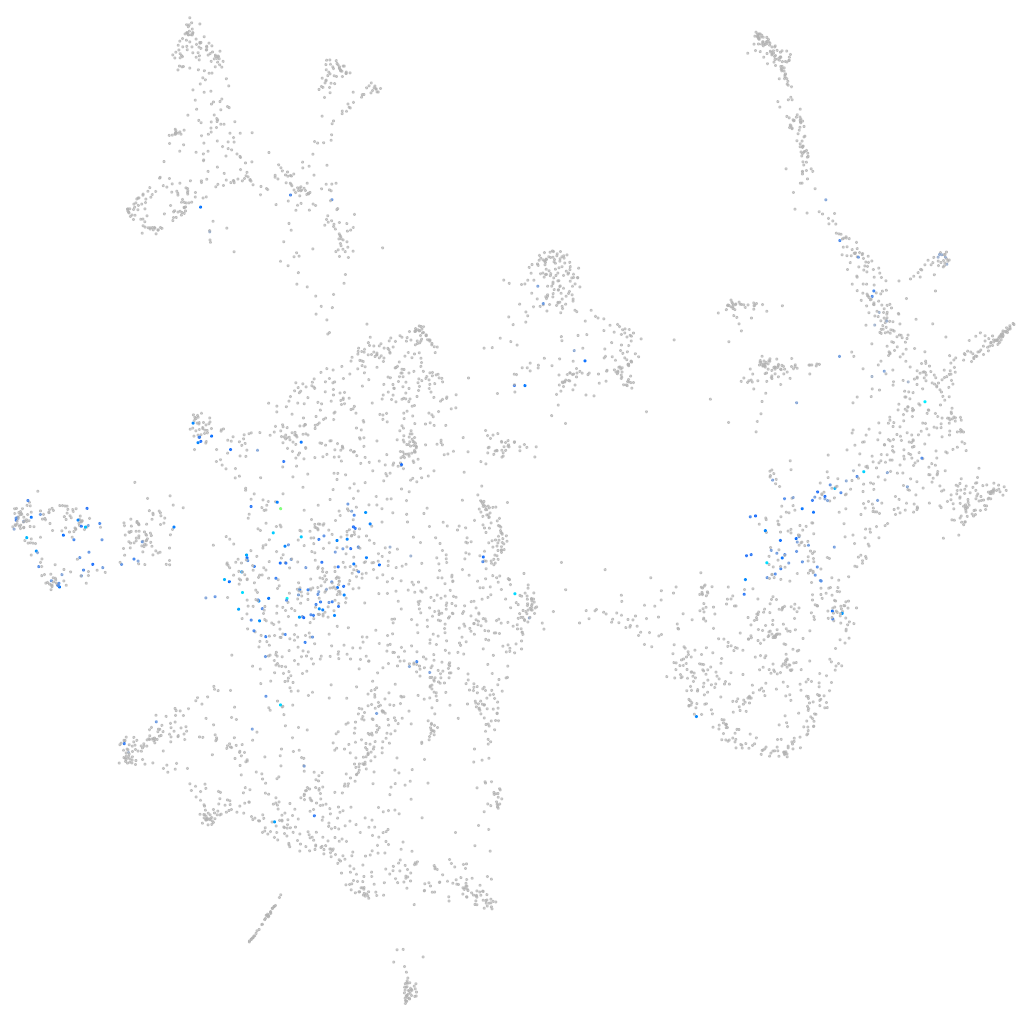
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:110216 | 0.374 | tob1b | -0.123 |
| zgc:165555.3 | 0.350 | gapdhs | -0.097 |
| zgc:110425 | 0.344 | tmem59 | -0.094 |
| ccna2 | 0.329 | ccni | -0.093 |
| mibp | 0.324 | pvalb1 | -0.092 |
| mki67 | 0.323 | pvalb2 | -0.091 |
| si:dkey-261m9.17 | 0.314 | nupr1a | -0.091 |
| cdca5 | 0.313 | gabarapa | -0.089 |
| cks1b | 0.301 | b2ml | -0.088 |
| LOC100148591 | 0.300 | rnasekb | -0.088 |
| fbxo5 | 0.294 | pvalb8 | -0.086 |
| rrm1 | 0.293 | COX3 | -0.086 |
| si:dkey-108k21.10 | 0.290 | eno1a | -0.082 |
| cdk1 | 0.283 | socs3a | -0.082 |
| dek | 0.279 | atp2b1a | -0.080 |
| smc4 | 0.274 | map1lc3a | -0.077 |
| aurkb | 0.273 | dedd1 | -0.077 |
| si:ch211-113a14.18 | 0.273 | si:dkey-16p21.8 | -0.076 |
| mad2l1 | 0.267 | calml4a | -0.075 |
| plk1 | 0.266 | gabarapb | -0.075 |
| hist1h4l | 0.264 | pkig | -0.074 |
| tpx2 | 0.263 | otofb | -0.073 |
| lbr | 0.262 | ddit3 | -0.073 |
| LOC100330864 | 0.261 | gabarapl2 | -0.073 |
| ncapd2 | 0.261 | nptna | -0.073 |
| dut | 0.260 | calm1a | -0.073 |
| chaf1a | 0.258 | cebpd | -0.073 |
| si:ch1073-153i20.4 | 0.258 | gpx2 | -0.073 |
| top2a | 0.256 | atp6v1e1b | -0.072 |
| ncapg | 0.255 | pgk1 | -0.072 |
| pcna | 0.254 | spint2 | -0.072 |
| rrm2 | 0.252 | vamp2 | -0.072 |
| g2e3 | 0.251 | actc1b | -0.071 |
| si:ch211-113a14.24 | 0.249 | creb3l3l | -0.071 |
| hmgb2a | 0.248 | mt-co2 | -0.071 |