"HIG1 hypoxia inducible domain family, member 2A"
ZFIN 
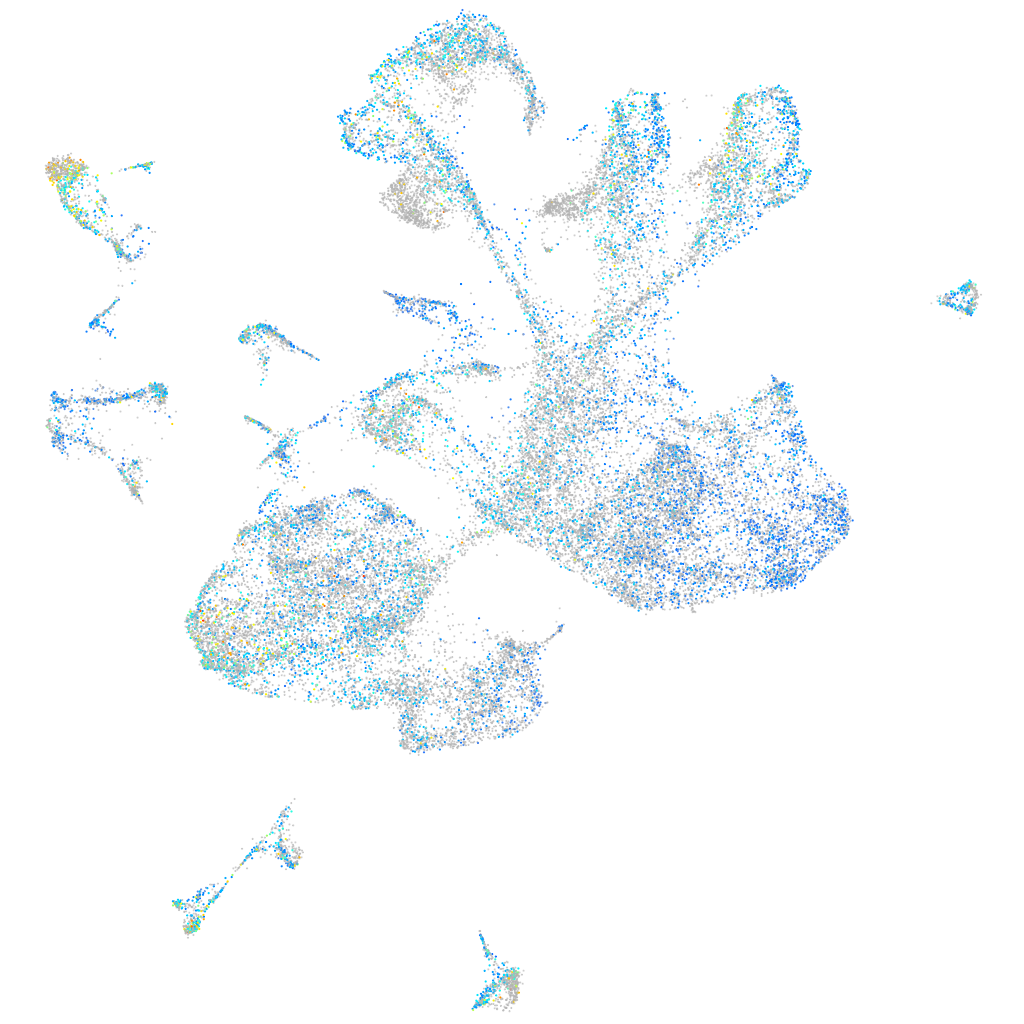
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| atp6v1g1 | 0.164 | XLOC-003689 | -0.101 |
| atp6v1e1b | 0.158 | stmn1a | -0.097 |
| tpi1b | 0.154 | hmgb2a | -0.095 |
| cdc42 | 0.152 | her15.1 | -0.093 |
| rnasekb | 0.150 | hmgb2b | -0.092 |
| necap1 | 0.149 | dla | -0.090 |
| cotl1 | 0.148 | si:dkey-151g10.6 | -0.089 |
| gapdhs | 0.146 | hmga1a | -0.089 |
| mllt11 | 0.144 | rplp1 | -0.088 |
| atp5mc1 | 0.144 | si:ch211-222l21.1 | -0.087 |
| acbd7 | 0.142 | XLOC-003692 | -0.083 |
| anxa13l | 0.141 | abhd6a | -0.083 |
| calm1a | 0.138 | si:ch73-21g5.7 | -0.083 |
| arpc3 | 0.137 | cldn5a | -0.082 |
| atp5if1b | 0.135 | notch1a | -0.081 |
| brk1 | 0.135 | slc1a3a | -0.079 |
| ywhag2 | 0.134 | mir219-3 | -0.078 |
| calm2a | 0.133 | XLOC-003690 | -0.078 |
| vdac3 | 0.132 | sb:cb81 | -0.077 |
| zgc:65894 | 0.132 | LOC798783 | -0.077 |
| gabarapl2 | 0.132 | hmgn2 | -0.076 |
| cox7a2a | 0.131 | lfng | -0.076 |
| cldnk | 0.130 | sox11b | -0.074 |
| anxa13 | 0.129 | si:ch211-57n23.4 | -0.073 |
| cox4i1 | 0.129 | cx43.4 | -0.073 |
| cd59 | 0.128 | si:dkey-42i9.4 | -0.073 |
| ndufa4 | 0.128 | rps29 | -0.072 |
| atp6ap2 | 0.128 | rplp2l | -0.072 |
| vat1 | 0.128 | lbr | -0.071 |
| rab6bb | 0.128 | notch1b | -0.071 |
| myl6 | 0.127 | notch3 | -0.071 |
| gng3 | 0.127 | her4.2 | -0.070 |
| atp6v0cb | 0.127 | her12 | -0.070 |
| ap2m1a | 0.126 | lrrn1 | -0.069 |
| mid1ip1b | 0.126 | nrarpb | -0.069 |