"HERV-H LTR-associating 2a, tandem duplicate 2"
ZFIN 
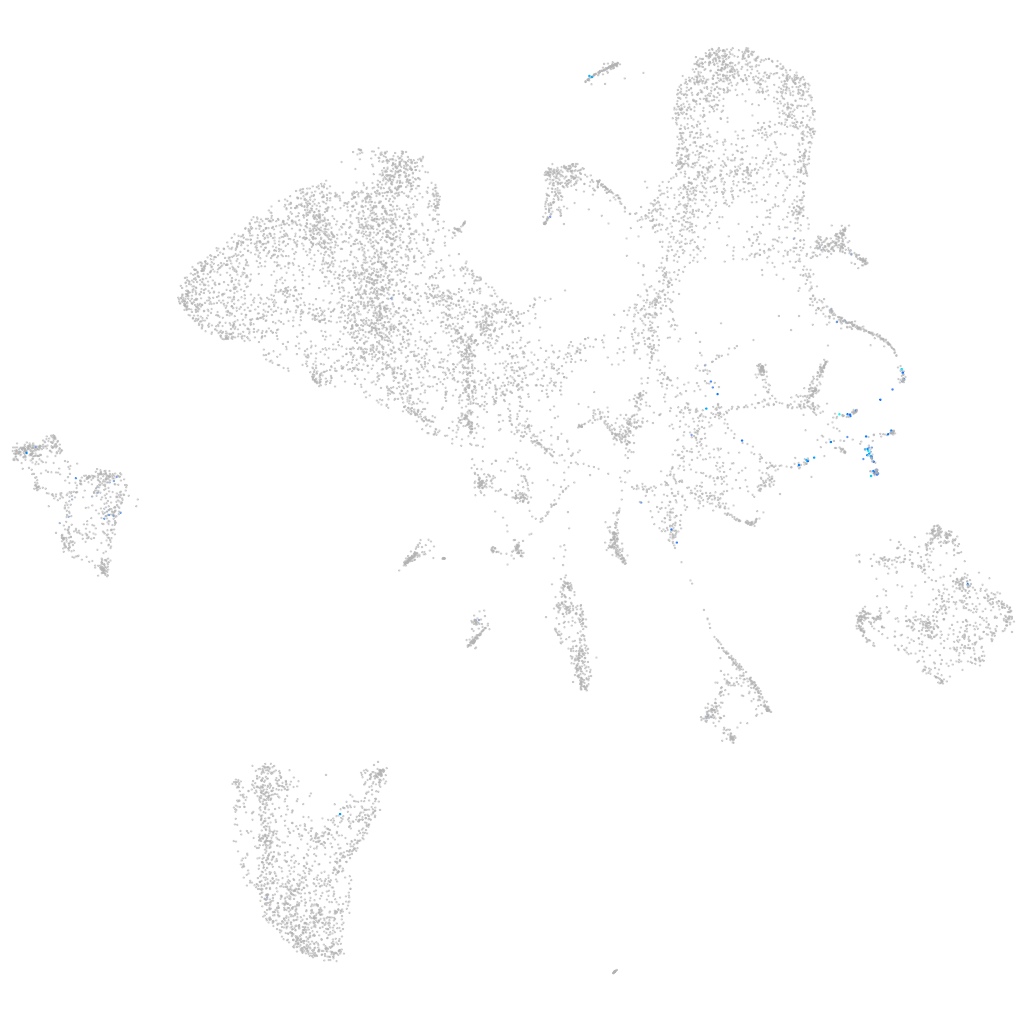
Other cell groups


all cells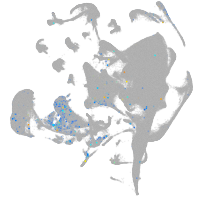 |
blastomeres |
PGCs |
endoderm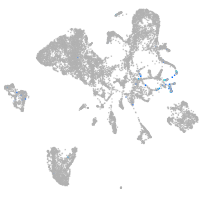 |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle 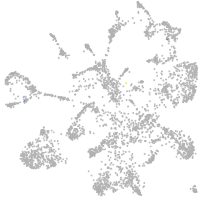 |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis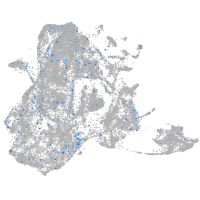 |
periderm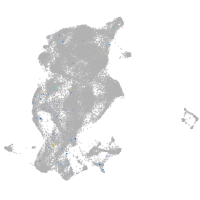 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| SLC5A10 | 0.377 | gapdh | -0.096 |
| ucn3l | 0.317 | gamt | -0.083 |
| ndufa4l2a | 0.305 | ahcy | -0.083 |
| sst1.1 | 0.288 | aldob | -0.078 |
| dkk3b | 0.271 | gatm | -0.075 |
| ptn | 0.268 | fbp1b | -0.073 |
| scg2a | 0.267 | apoc2 | -0.070 |
| slc30a2 | 0.262 | apoa4b.1 | -0.070 |
| pcsk2 | 0.260 | glud1b | -0.069 |
| srd5a2b | 0.252 | apoa1b | -0.069 |
| kcnj11 | 0.249 | bhmt | -0.069 |
| rims2a | 0.243 | mat1a | -0.067 |
| pcsk1 | 0.243 | afp4 | -0.067 |
| camk1da | 0.242 | gpx4a | -0.066 |
| gnrh2 | 0.236 | gstt1a | -0.066 |
| cntnap2a | 0.235 | cx32.3 | -0.065 |
| scgn | 0.234 | nupr1b | -0.063 |
| prokr1a | 0.228 | scp2a | -0.062 |
| shisal1a | 0.227 | si:dkey-16p21.8 | -0.062 |
| fam49bb | 0.226 | sod1 | -0.061 |
| g6pcb | 0.220 | apoa2 | -0.061 |
| si:dkey-153k10.9 | 0.219 | aqp12 | -0.061 |
| cadm2a | 0.217 | eno3 | -0.060 |
| LOC571123 | 0.216 | cdo1 | -0.060 |
| cpne4a | 0.216 | gstr | -0.060 |
| tspan7b | 0.214 | sult2st2 | -0.059 |
| prlra | 0.210 | agxta | -0.059 |
| tmem163b | 0.209 | abat | -0.059 |
| scg2b | 0.208 | suclg1 | -0.058 |
| CR774179.5 | 0.207 | chchd10 | -0.058 |
| ins | 0.205 | gcshb | -0.058 |
| BX927253.3 | 0.205 | apobb.1 | -0.058 |
| drd5a | 0.203 | suclg2 | -0.058 |
| gpr22a | 0.203 | dhrs9 | -0.058 |
| kcnk9 | 0.203 | hspe1 | -0.057 |