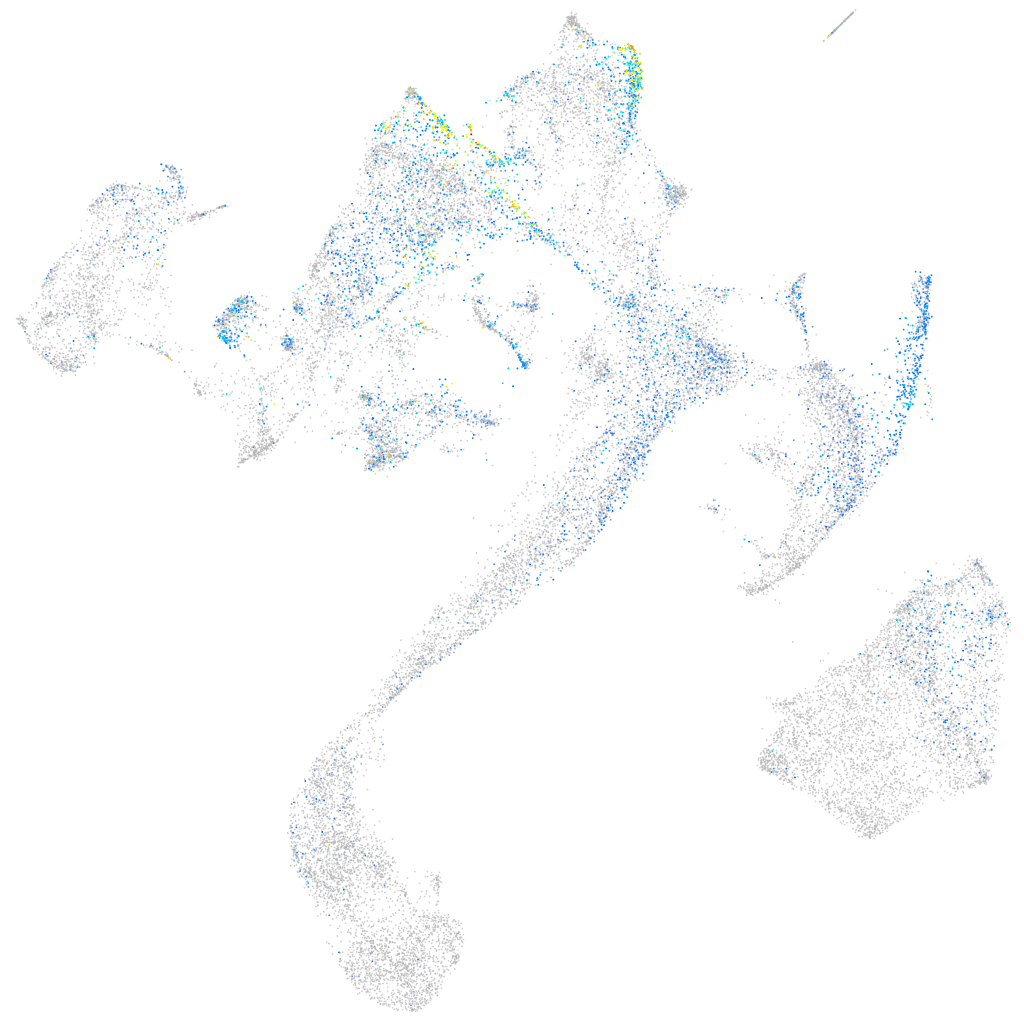
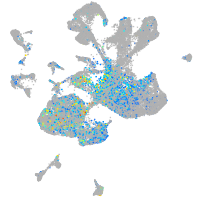
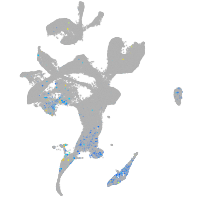
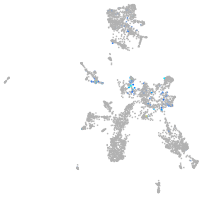
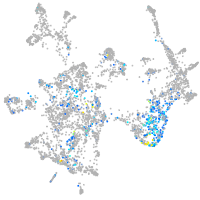
hedgehog interacting protein
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| krt18a.1 | 0.288 | ckmb | -0.150 |
| krt8 | 0.276 | atp2a1 | -0.149 |
| en1b | 0.274 | gapdh | -0.148 |
| ptch2 | 0.265 | ckma | -0.148 |
| en1a | 0.259 | ak1 | -0.138 |
| col5a1 | 0.256 | si:ch73-367p23.2 | -0.138 |
| fstl1b | 0.254 | aldoab | -0.137 |
| mfap2 | 0.252 | ank1a | -0.137 |
| marcksl1a | 0.248 | hspb1 | -0.136 |
| col1a1b | 0.245 | ldb3b | -0.134 |
| col5a2a | 0.244 | cycsb | -0.133 |
| itgbl1 | 0.243 | tmem38a | -0.133 |
| col1a2 | 0.242 | actn3a | -0.131 |
| sparc | 0.236 | myom1a | -0.131 |
| dcn | 0.230 | tnnc2 | -0.130 |
| zgc:153867 | 0.230 | pgam2 | -0.130 |
| fbn2b | 0.230 | tnnt3a | -0.130 |
| tgfbi | 0.229 | hhatla | -0.130 |
| ppib | 0.229 | neb | -0.129 |
| si:ch1073-459j12.1 | 0.228 | mylz3 | -0.127 |
| vwde | 0.224 | prx | -0.127 |
| fkbp14 | 0.224 | myl1 | -0.127 |
| pdgfrl | 0.223 | acta1b | -0.126 |
| col1a1a | 0.222 | mylpfa | -0.126 |
| nkx3.2 | 0.221 | pkmb | -0.125 |
| fstl1a | 0.218 | casq1b | -0.125 |
| cxcl12a | 0.218 | smyd1a | -0.125 |
| pdia3 | 0.217 | pabpc4 | -0.124 |
| clu | 0.216 | si:ch211-266g18.10 | -0.124 |
| actb2 | 0.216 | srl | -0.122 |
| pmp22a | 0.216 | nme2b.2 | -0.121 |
| ssr3 | 0.214 | myom2a | -0.121 |
| col8a1a | 0.213 | XLOC-001975 | -0.121 |
| lrrc17 | 0.212 | eef2l2 | -0.121 |
| ckap4 | 0.211 | XLOC-025819 | -0.121 |