HECT and RLD domain containing E3 ubiquitin protein ligase 2
ZFIN 
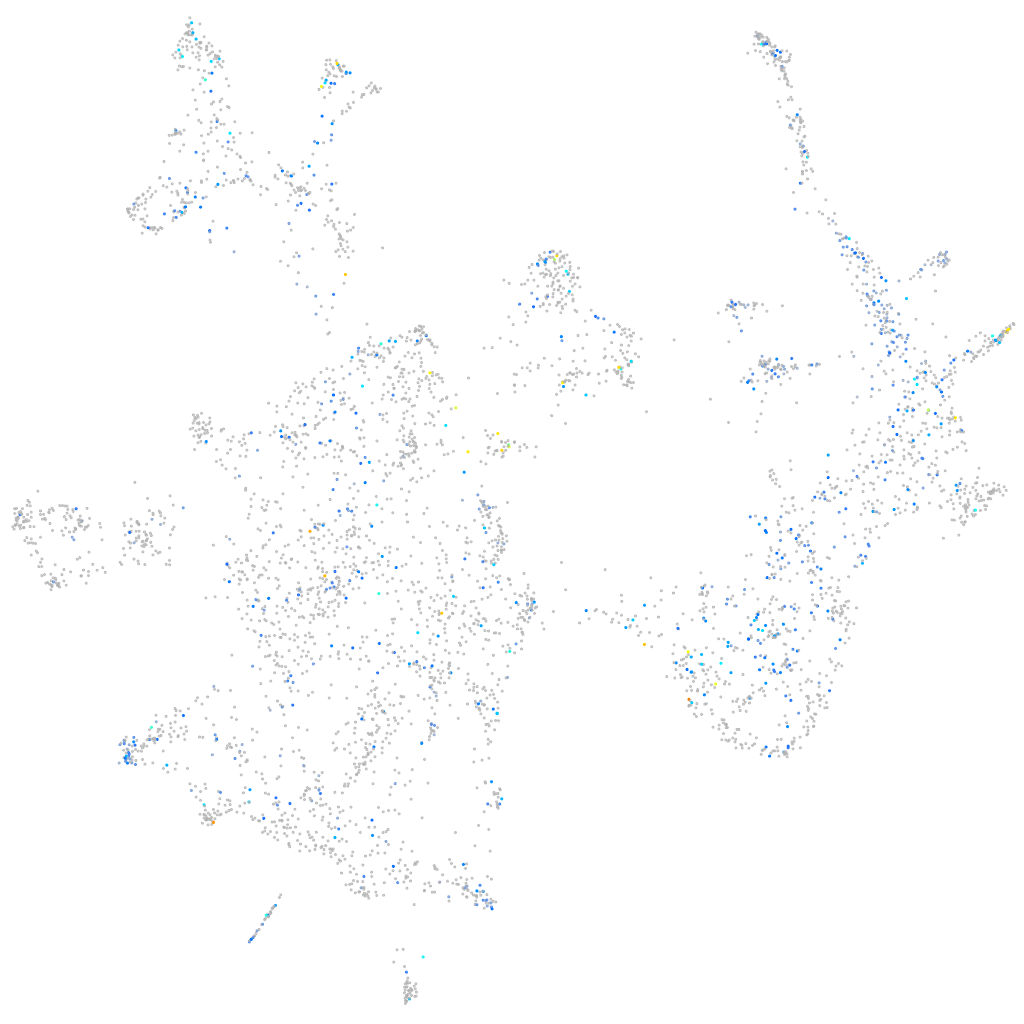
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| LOC101886946.1 | 0.119 | FP017215.1 | -0.088 |
| notchl | 0.115 | cldna | -0.078 |
| XLOC-029934 | 0.114 | agr2 | -0.076 |
| mast1a | 0.111 | bricd5 | -0.070 |
| slain1b | 0.110 | mdka | -0.068 |
| uts2d | 0.109 | zbtb16a | -0.066 |
| chst12b | 0.109 | zgc:194210 | -0.065 |
| urp1 | 0.108 | oc90 | -0.063 |
| zgc:123107 | 0.107 | wfdc2 | -0.062 |
| LOC103911601 | 0.107 | rps4x | -0.060 |
| LOC110439513 | 0.106 | wu:fc01d11 | -0.060 |
| LOC101882339 | 0.106 | crtap | -0.060 |
| LOC110437817 | 0.104 | rps27.1 | -0.059 |
| krt18b | 0.104 | eif4a3 | -0.058 |
| gucy1a1 | 0.102 | pkdcca | -0.057 |
| XLOC-036507 | 0.101 | btg3 | -0.057 |
| nnr | 0.101 | LOC100331480 | -0.056 |
| XLOC-001298 | 0.100 | rps5 | -0.056 |
| CABZ01086574.1 | 0.100 | tpm4a | -0.055 |
| XLOC-033543 | 0.099 | efhd1 | -0.055 |
| she | 0.098 | rps2 | -0.055 |
| btr01 | 0.096 | prdm12b | -0.055 |
| mlycd | 0.095 | stm | -0.055 |
| kcnj2b | 0.095 | hmgb3b | -0.055 |
| slc1a7b | 0.095 | ccnd1 | -0.054 |
| kremen1 | 0.095 | cdh11 | -0.054 |
| gpr185b | 0.095 | col2a1b | -0.054 |
| sema3ga | 0.095 | col9a1b | -0.054 |
| BX248238.1 | 0.095 | ucmab | -0.053 |
| LOC108191590 | 0.095 | sumo3a | -0.053 |
| CR762407.1 | 0.094 | sumo3b | -0.053 |
| nipsnap3a | 0.093 | atic | -0.053 |
| sox1a | 0.092 | rpl23 | -0.053 |
| cd109 | 0.091 | eef1b2 | -0.052 |
| LOC110440047 | 0.091 | eva1a | -0.052 |