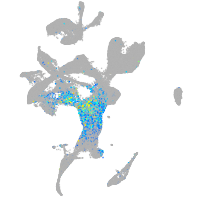
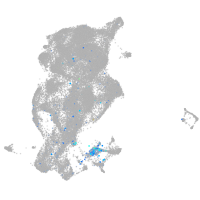
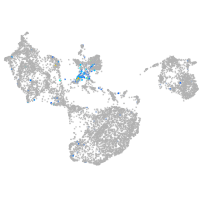
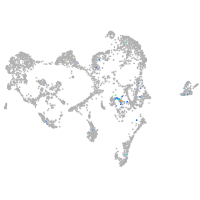
"hairy-related 4, tandem duplicate 4"
ZFIN 
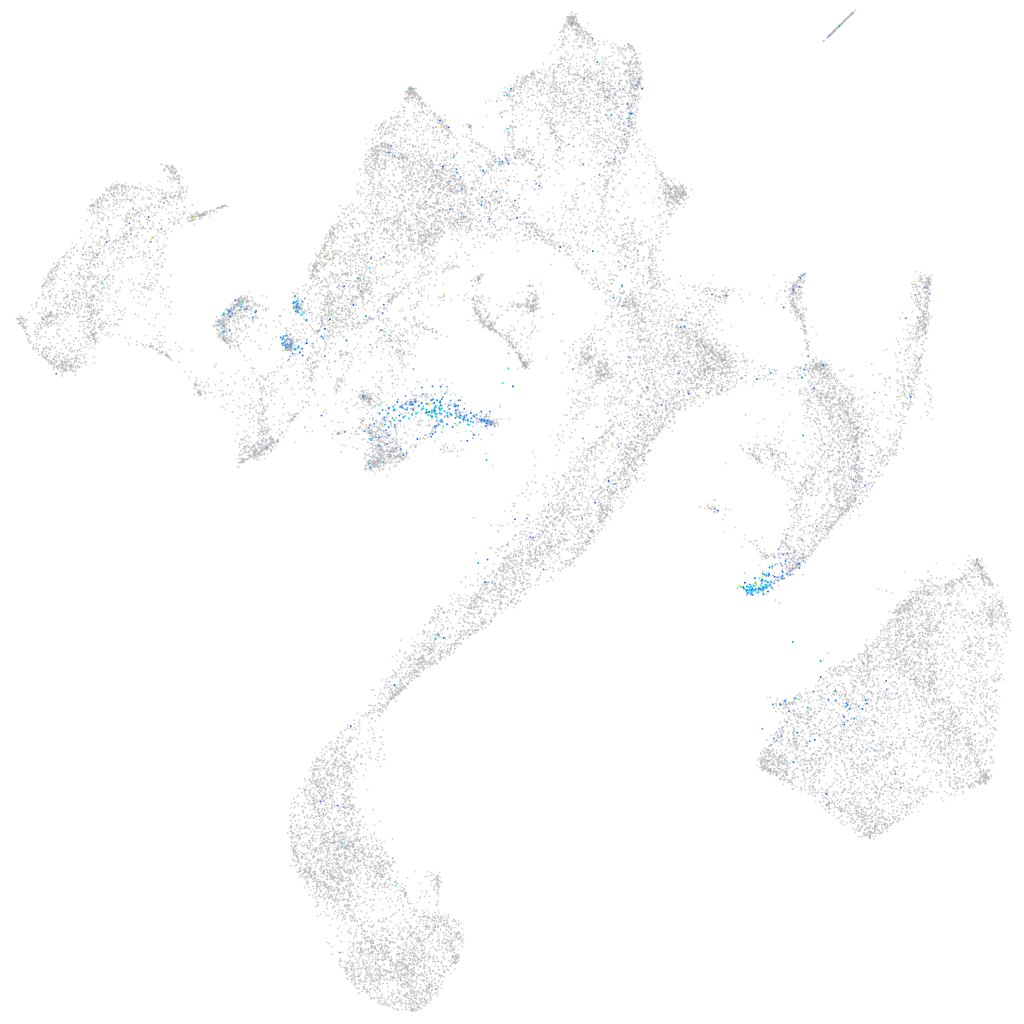
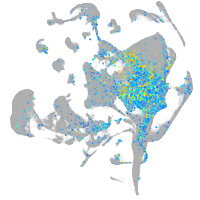
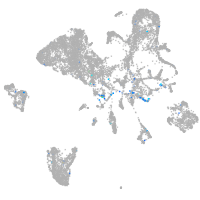
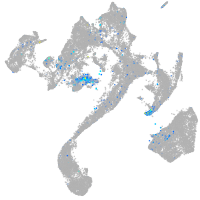
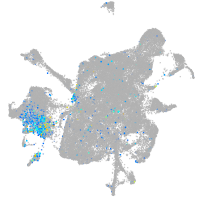
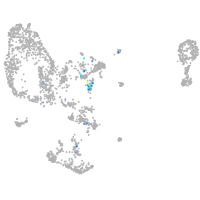
Expression by stage/cluster


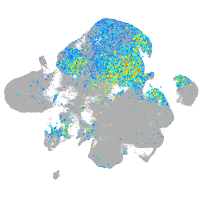
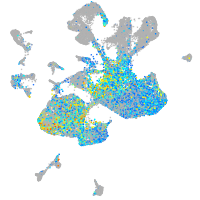
Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| her4.2 | 0.514 | tuba8l2 | -0.075 |
| her4.1 | 0.414 | ttn.1 | -0.063 |
| XLOC-003689 | 0.377 | tnnc2 | -0.058 |
| XLOC-003690 | 0.375 | neb | -0.057 |
| XLOC-003692 | 0.374 | ldb3b | -0.057 |
| her15.1 | 0.366 | actn3b | -0.057 |
| abhd6a | 0.362 | si:ch211-266g18.10 | -0.057 |
| her2 | 0.252 | gapdh | -0.057 |
| si:ch73-21g5.7 | 0.244 | actn3a | -0.055 |
| LOC798783 | 0.230 | myom1a | -0.055 |
| her12 | 0.199 | si:ch73-367p23.2 | -0.055 |
| zgc:165461 | 0.198 | ak1 | -0.055 |
| dla | 0.197 | aldoab | -0.055 |
| gadd45gb.1 | 0.194 | ttn.2 | -0.055 |
| CU467822.1 | 0.192 | ldb3a | -0.054 |
| dlb | 0.185 | tmod4 | -0.054 |
| cldn5a | 0.184 | smyd1a | -0.054 |
| notch3 | 0.177 | cav3 | -0.054 |
| CU634008.1 | 0.176 | atp2a1 | -0.054 |
| her4.3 | 0.171 | pgam2 | -0.053 |
| nova2 | 0.165 | nme2b.2 | -0.052 |
| msi1 | 0.161 | mybphb | -0.052 |
| tuba1c | 0.156 | pabpc4 | -0.052 |
| insm1a | 0.154 | ckmb | -0.052 |
| elavl3 | 0.152 | casq1b | -0.052 |
| her9 | 0.149 | hhatla | -0.051 |
| ptmaa | 0.148 | eef2l2 | -0.051 |
| fabp7a | 0.148 | si:dkey-16p21.8 | -0.051 |
| pou3f3b | 0.146 | ckma | -0.051 |
| tuba1a | 0.146 | tpma | -0.050 |
| her13 | 0.141 | XLOC-001975 | -0.050 |
| gfap | 0.139 | prx | -0.050 |
| mir219-3 | 0.138 | srl | -0.050 |
| sox2 | 0.138 | ank1a | -0.050 |
| neurod4 | 0.135 | myom2a | -0.050 |