hydroxyacyl-CoA dehydrogenase trifunctional multienzyme complex subunit alpha b
ZFIN 
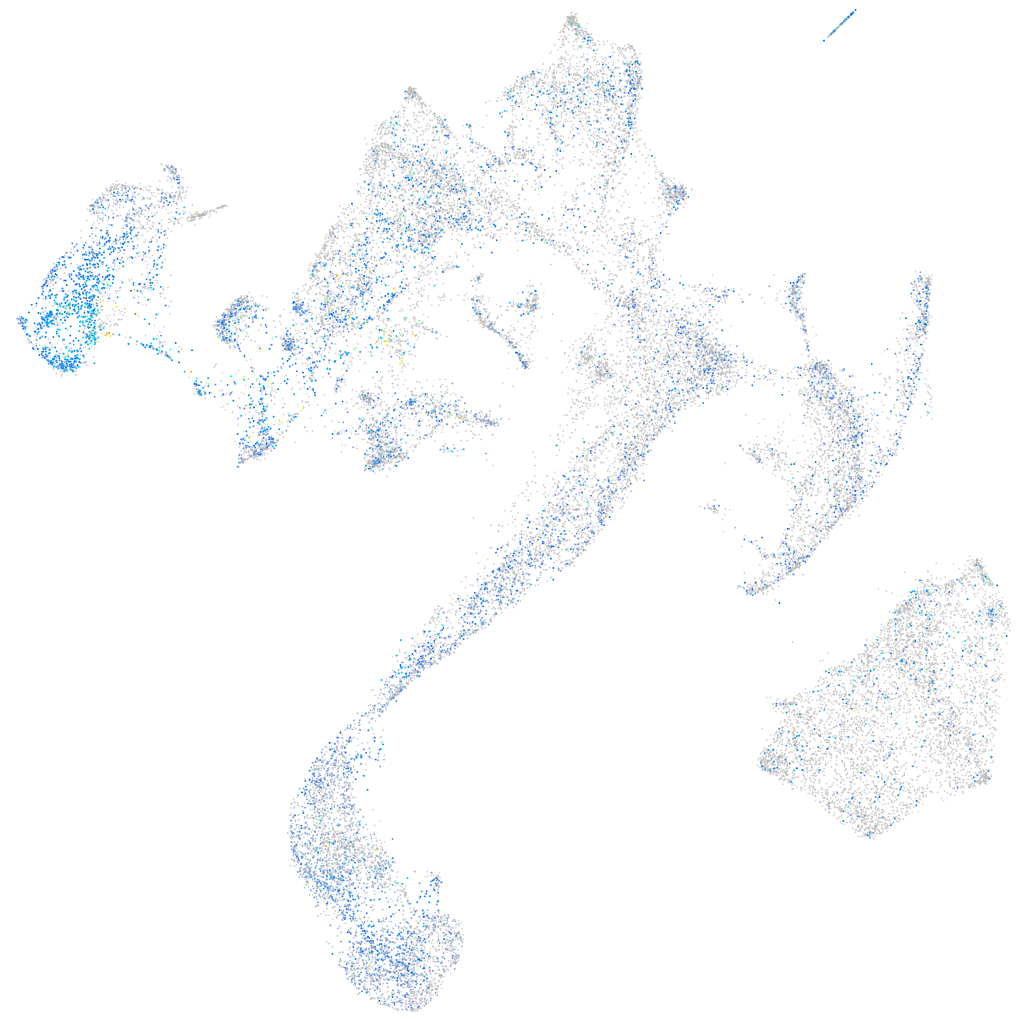
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| tnnt2e | 0.321 | hsp90ab1 | -0.245 |
| got1 | 0.317 | h3f3d | -0.232 |
| tnnt2d | 0.316 | eef1a1l1 | -0.230 |
| pvalb4 | 0.313 | si:ch73-1a9.3 | -0.230 |
| mybpc3 | 0.312 | pabpc1a | -0.229 |
| tnni4b.2 | 0.306 | cirbpb | -0.222 |
| aco2 | 0.305 | si:ch73-281n10.2 | -0.213 |
| tnnc1b | 0.304 | khdrbs1a | -0.208 |
| mt-atp6 | 0.304 | h2afvb | -0.208 |
| BX248497.1 | 0.302 | ran | -0.205 |
| nnt | 0.301 | si:ch211-222l21.1 | -0.205 |
| hhatlb | 0.301 | hmgb2b | -0.203 |
| atp5if1b | 0.300 | fthl27 | -0.202 |
| ugp2a | 0.296 | hnrnpaba | -0.201 |
| atp5mc1 | 0.294 | hmgb2a | -0.201 |
| LO018250.1 | 0.294 | hmga1a | -0.196 |
| chchd10 | 0.291 | naca | -0.195 |
| mdh2 | 0.289 | ptmab | -0.195 |
| mt-nd4 | 0.289 | hnrnpabb | -0.191 |
| prx | 0.289 | cx43.4 | -0.189 |
| mt-nd2 | 0.284 | syncrip | -0.189 |
| atp5f1b | 0.283 | cbx3a | -0.189 |
| sptb | 0.283 | ubc | -0.184 |
| eno3 | 0.283 | anp32a | -0.184 |
| cs | 0.282 | hnrnpa0b | -0.183 |
| glud1b | 0.282 | anp32b | -0.183 |
| ryr1a | 0.281 | hmgn6 | -0.180 |
| casq2 | 0.281 | nucks1a | -0.179 |
| gapdh | 0.281 | hmgn7 | -0.178 |
| pygmb | 0.280 | cirbpa | -0.178 |
| zgc:154046 | 0.280 | setb | -0.177 |
| atp5meb | 0.280 | nop58 | -0.174 |
| myom1b | 0.280 | seta | -0.173 |
| sucla2 | 0.278 | snrpb | -0.172 |
| atp5po | 0.278 | rbm8a | -0.172 |