"H1 histone family, member 0 [Source:ZFIN;Acc:ZDB-GENE-030131-337]"
ZFIN 
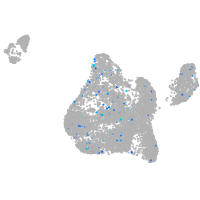
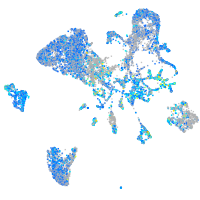
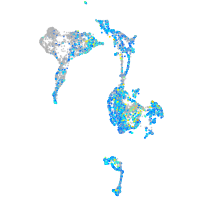
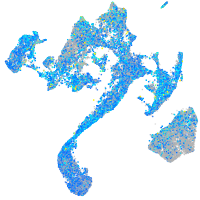
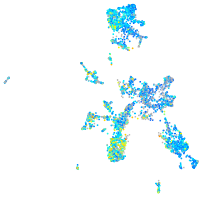
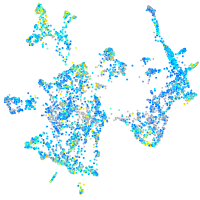
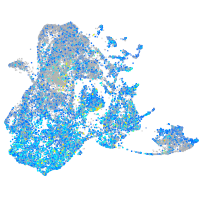
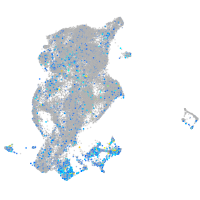
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR383676.1 | 0.263 | gapdh | -0.224 |
| epcam | 0.263 | ahcy | -0.202 |
| histh1l | 0.257 | gstt1a | -0.200 |
| gapdhs | 0.255 | apoc2 | -0.199 |
| cldn7b | 0.247 | gamt | -0.185 |
| ier2b | 0.247 | scp2a | -0.183 |
| jun | 0.241 | hspe1 | -0.182 |
| cldnb | 0.236 | si:dkey-16p21.8 | -0.181 |
| scg3 | 0.233 | apoa4b.1 | -0.181 |
| h1fx | 0.233 | sult2st2 | -0.177 |
| spint2 | 0.228 | fbp1b | -0.176 |
| ccni | 0.227 | apoa1b | -0.174 |
| jpt1b | 0.223 | mat1a | -0.170 |
| neurod1 | 0.223 | gcshb | -0.168 |
| fosab | 0.223 | msrb2 | -0.166 |
| btg2 | 0.223 | chchd10 | -0.164 |
| calm1b | 0.221 | nipsnap3a | -0.164 |
| txnipa | 0.219 | gatm | -0.163 |
| foxp4 | 0.217 | afp4 | -0.162 |
| tmem59 | 0.217 | apoc1 | -0.160 |
| cldnh | 0.217 | si:ch211-201h21.5 | -0.157 |
| si:ch1073-429i10.3.1 | 0.215 | eno3 | -0.157 |
| marcksl1a | 0.214 | ftcd | -0.156 |
| cd9b | 0.214 | gpx4a | -0.156 |
| btg1 | 0.214 | hspd1 | -0.155 |
| mir375-2 | 0.213 | aldob | -0.154 |
| tox | 0.212 | suclg1 | -0.154 |
| anxa4 | 0.211 | cyp8b1 | -0.148 |
| vat1 | 0.207 | bhmt | -0.146 |
| calm1a | 0.207 | phyhd1 | -0.144 |
| zgc:92380 | 0.207 | gstr | -0.140 |
| atf4b | 0.206 | ugt1a7 | -0.140 |
| si:ch211-196l7.4 | 0.205 | agxta | -0.139 |
| gnb1a | 0.205 | npm1a | -0.138 |
| f11r.1 | 0.205 | rdh1 | -0.138 |