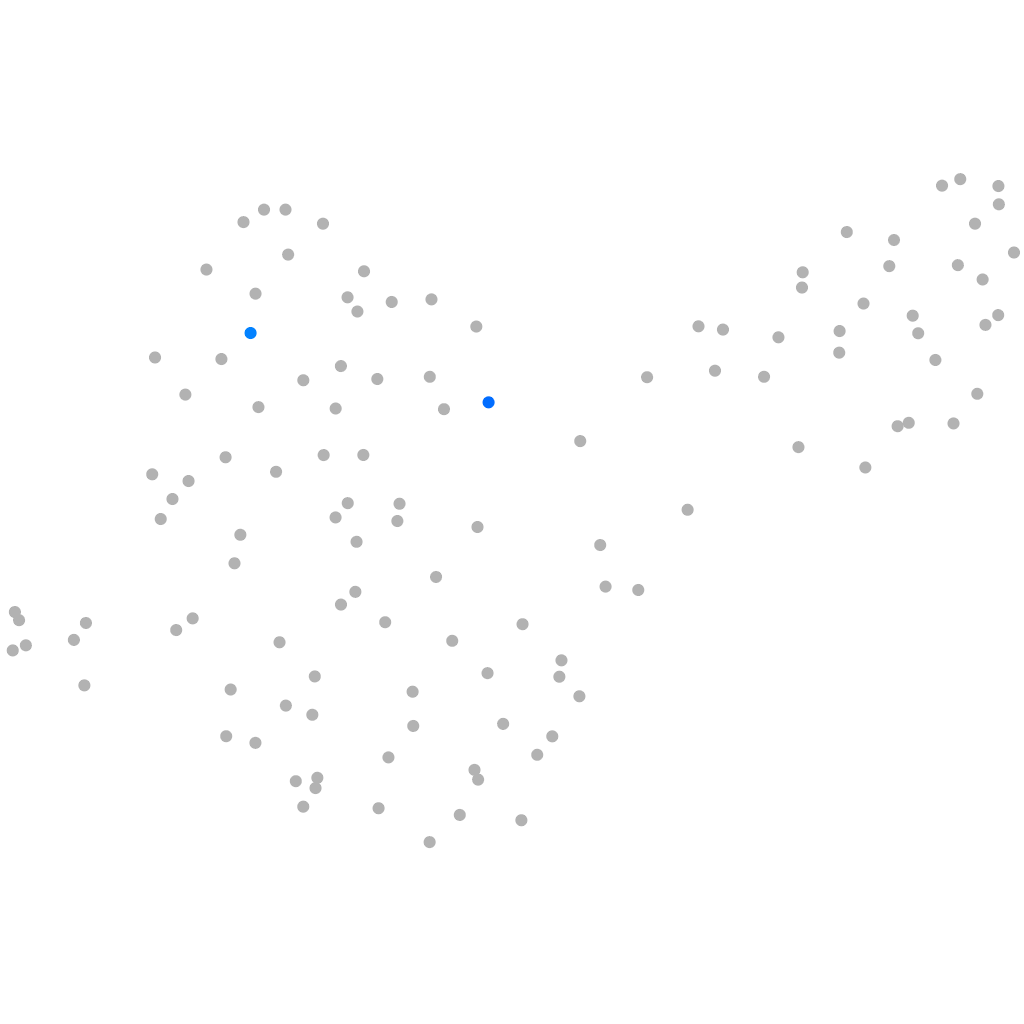
granzyme K
ZFIN 
Other cell groups


all cells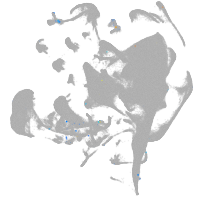 |
blastomeres |
PGCs |
endoderm |
axial mesoderm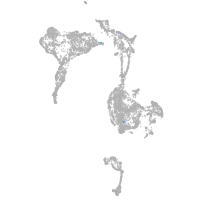 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 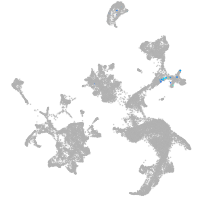 |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory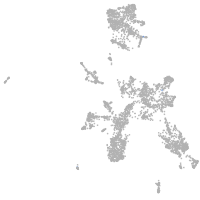 |
otic / lateral line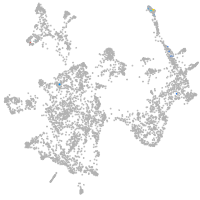 |
epidermis |
periderm |
fin |
ionocytes / mucous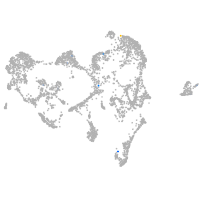 |
pigment cells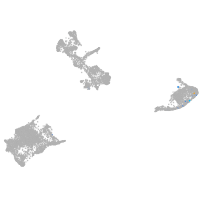 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| adra1ab | 1.000 | hnrnpaba | -0.272 |
| si:dkeyp-72e1.7 | 1.000 | npc2 | -0.241 |
| unc45b | 0.850 | hnrnpa0b | -0.238 |
| lmbrd2a | 0.805 | ilf3b | -0.237 |
| prkchb | 0.797 | actb1 | -0.228 |
| ftr97 | 0.777 | sec61b | -0.219 |
| CT737210.1 | 0.763 | hmga1a | -0.208 |
| ACKR2 | 0.763 | anp32e | -0.203 |
| adarb1b | 0.763 | ptmab | -0.201 |
| AL590150.1 | 0.763 | snrpa | -0.200 |
| AL929304.1 | 0.763 | actb2 | -0.200 |
| arhgef3 | 0.763 | ranbp1 | -0.195 |
| bsna | 0.763 | h1m | -0.190 |
| BX323991.1 | 0.763 | ccna2 | -0.185 |
| BX470229.2 | 0.763 | sltm | -0.184 |
| BX510909.2 | 0.763 | ybx1 | -0.183 |
| BX530017.2 | 0.763 | faf2 | -0.183 |
| BX530024.1 | 0.763 | banf1 | -0.181 |
| BX927308.1 | 0.763 | rbm7 | -0.181 |
| cd247l | 0.763 | dnajc1 | -0.180 |
| cdc42se2 | 0.763 | hnrnpl | -0.179 |
| CR589875.1 | 0.763 | rbm4.3 | -0.175 |
| crybb2 | 0.763 | odc1 | -0.172 |
| diras1b | 0.763 | nucks1a | -0.172 |
| dkk1a | 0.763 | spcs2 | -0.171 |
| dock11 | 0.763 | skp1 | -0.169 |
| fam151a | 0.763 | srsf3b | -0.169 |
| FO834888.1 | 0.763 | ctsd | -0.167 |
| gabbr1a | 0.763 | rhoac | -0.167 |
| glra1 | 0.763 | AL935174.5 | -0.167 |
| gramd4a | 0.763 | atf7ip | -0.165 |
| helt | 0.763 | hspb1 | -0.164 |
| il7r | 0.763 | smarca4a | -0.163 |
| kcnk13b | 0.763 | marcksb | -0.163 |
| LOC100331003 | 0.763 | gar1 | -0.163 |