"glucuronidase, beta"
ZFIN 
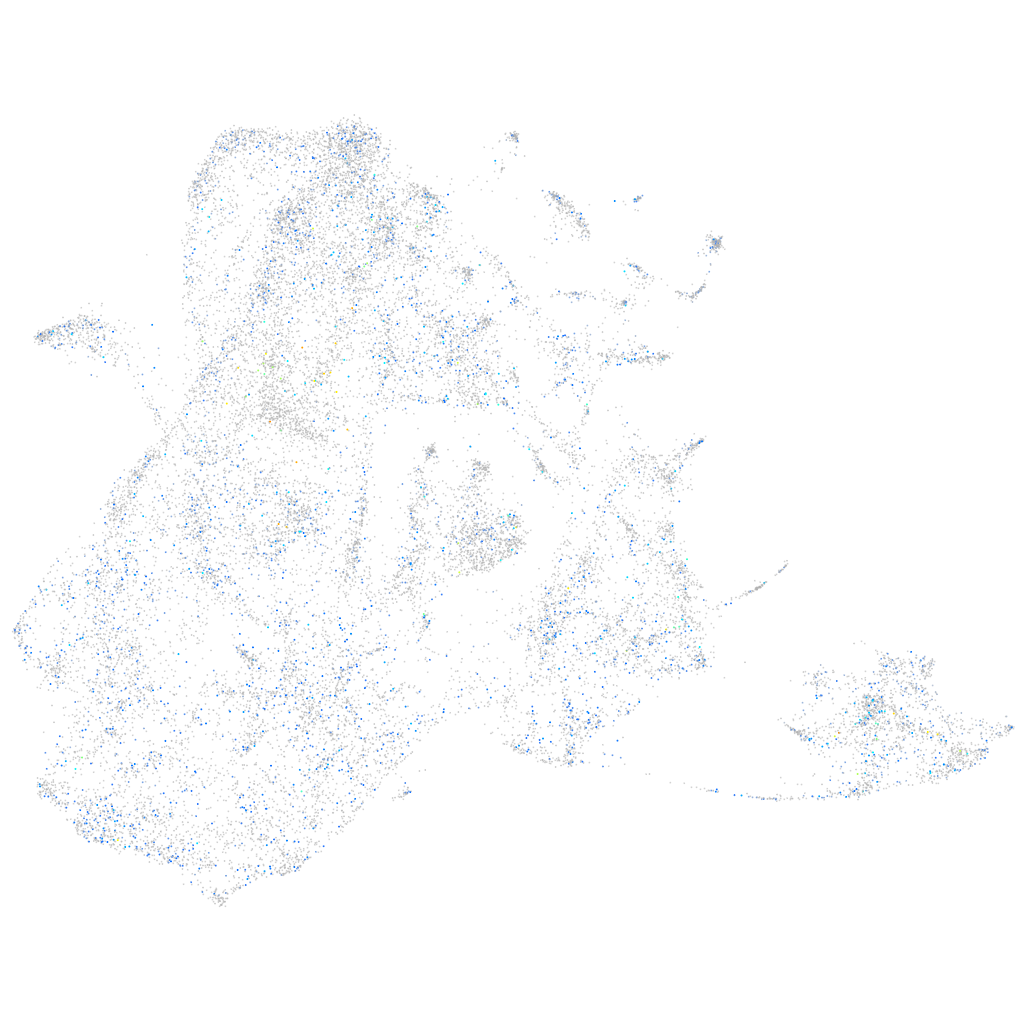
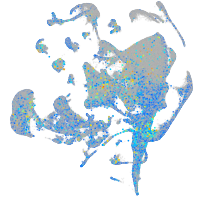
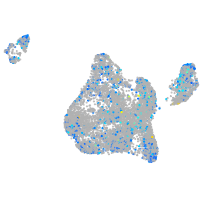
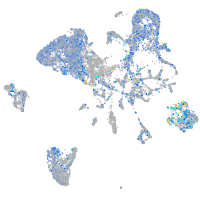
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| mt-cyb | 0.084 | actb2 | -0.051 |
| NC-002333.4 | 0.080 | lgals1l1 | -0.048 |
| mt-co2 | 0.080 | pfn1 | -0.048 |
| mt-atp6 | 0.075 | krt5 | -0.047 |
| mt-nd4 | 0.074 | cotl1 | -0.047 |
| COX3 | 0.072 | krtt1c19e | -0.046 |
| mt-nd1 | 0.070 | dap1b | -0.046 |
| CABZ01072614.1 | 0.066 | zgc:136930 | -0.045 |
| mt-nd2 | 0.064 | krt91 | -0.043 |
| mt-co1 | 0.062 | si:dkey-183i3.5 | -0.042 |
| galnt8b.2 | 0.061 | si:dkey-33c14.3 | -0.042 |
| NC-002333.17 | 0.061 | tmsb4x | -0.041 |
| LOC108179687 | 0.058 | tmsb1 | -0.041 |
| slc39a7 | 0.056 | rbp4 | -0.040 |
| khsrp | 0.056 | aldh9a1a.1 | -0.040 |
| nucks1a | 0.056 | rpl39 | -0.039 |
| CR450824.1 | 0.055 | cfl1l | -0.038 |
| BX324164.1 | 0.055 | aep1 | -0.038 |
| LOC110438679 | 0.054 | epgn | -0.037 |
| LOC103910812 | 0.053 | cldn1 | -0.036 |
| arid3c | 0.052 | zgc:86896 | -0.036 |
| seta | 0.052 | gapdhs | -0.036 |
| syncrip | 0.052 | si:dkey-16p21.8 | -0.036 |
| canx | 0.052 | sparc | -0.034 |
| acin1a | 0.052 | cxl34b.11 | -0.034 |
| srsf1a | 0.052 | wu:fa03e10 | -0.033 |
| tmed10 | 0.052 | olfml3b | -0.032 |
| mtdha | 0.051 | zgc:153867 | -0.032 |
| AL935186.6 | 0.051 | cap1 | -0.032 |
| si:ch211-264f5.6 | 0.051 | lgals3b | -0.031 |
| sh3bp2 | 0.051 | ppdpfb | -0.031 |
| mt-nd5 | 0.051 | mgarp | -0.031 |
| hnrnpa1b | 0.050 | romo1 | -0.030 |
| snrpb | 0.050 | cdo1 | -0.030 |
| fbn2b | 0.050 | zgc:175088 | -0.030 |