"guanylate cyclase 2D, retinal"
ZFIN 
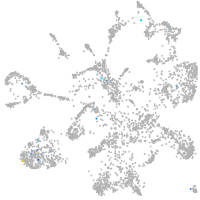
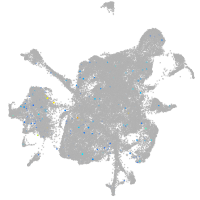
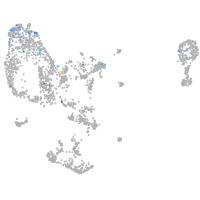
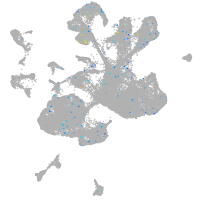
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| impg2a | 0.374 | gstp1 | -0.050 |
| prph2a | 0.340 | gch2 | -0.041 |
| guca1c | 0.318 | ahnak | -0.040 |
| gnat2 | 0.296 | slc2a15a | -0.037 |
| cnga3b | 0.283 | TMEM19 | -0.037 |
| tmx3a | 0.275 | actb2 | -0.036 |
| CR774179.2 | 0.266 | rab38 | -0.036 |
| si:ch1073-303d10.1 | 0.260 | prdx1 | -0.035 |
| mef2ab | 0.260 | si:dkey-251i10.2 | -0.035 |
| tulp1b | 0.249 | prdx6 | -0.034 |
| pde6c | 0.245 | C12orf75 | -0.032 |
| si:dkeyp-57d7.4 | 0.242 | krt18b | -0.032 |
| pdcb | 0.242 | pfn1 | -0.032 |
| cdhr1a | 0.239 | rab34b | -0.032 |
| zgc:73359 | 0.238 | slc2a11b | -0.031 |
| rgs9a | 0.235 | cmtm3 | -0.031 |
| grk1b | 0.235 | akr1b1 | -0.030 |
| si:ch211-203k16.3 | 0.232 | hbae3 | -0.030 |
| ckmt2a | 0.232 | bscl2l | -0.030 |
| prph2b | 0.231 | slc22a7a | -0.030 |
| guk1b | 0.231 | syngr1a | -0.030 |
| CR847998.1 | 0.228 | prps1a | -0.029 |
| guca1g | 0.227 | hbae1.1 | -0.029 |
| cnga3a | 0.227 | zgc:110339 | -0.029 |
| inaa | 0.221 | gpd1b | -0.029 |
| aipl2 | 0.219 | tuba8l3 | -0.028 |
| LOC101884966 | 0.214 | tmem243a | -0.028 |
| LOC100333290 | 0.213 | si:dkey-151g10.6 | -0.027 |
| rcvrn3 | 0.210 | dync1i2a | -0.027 |
| gnb3b | 0.210 | CABZ01021592.1 | -0.027 |
| rcvrn2 | 0.210 | cyb561a3b | -0.026 |
| zgc:195245 | 0.208 | cmtm7 | -0.026 |
| arr3b | 0.208 | si:ch211-194k22.8 | -0.026 |
| LOC110439885 | 0.205 | cax1 | -0.026 |
| impg1a | 0.202 | yipf4 | -0.026 |