glutathione S-transferase mu tandem duplicate 3
ZFIN 
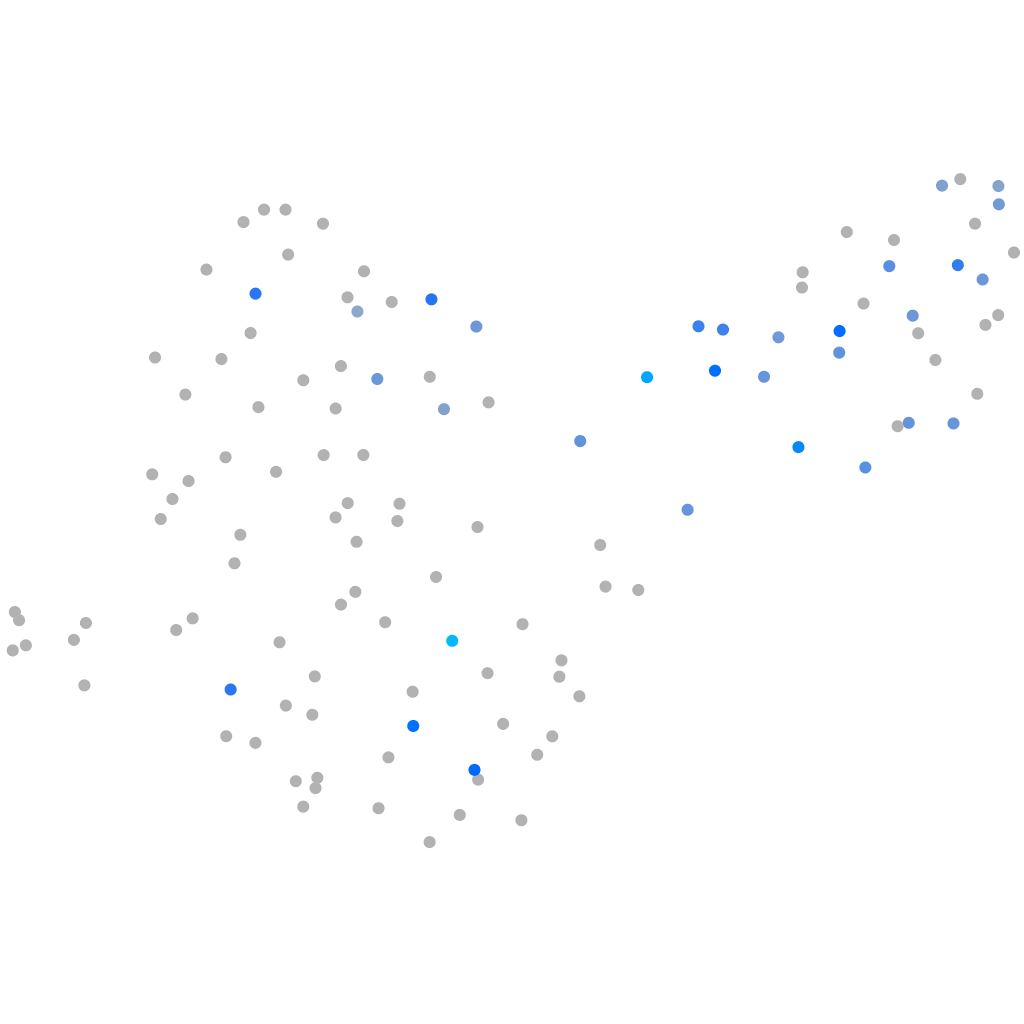
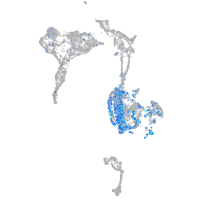
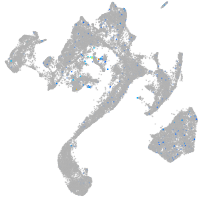
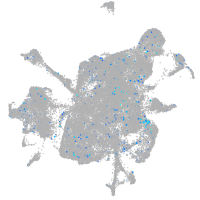
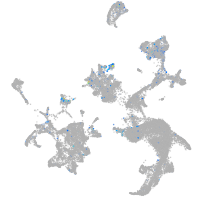
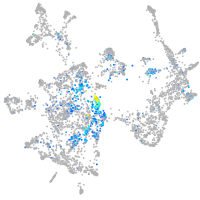
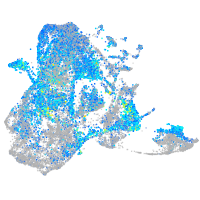
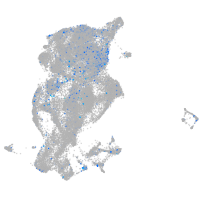
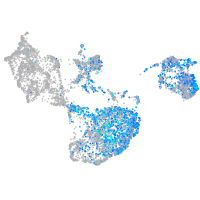
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cox4i2 | 0.599 | hnrnpa1b | -0.474 |
| ndufa3 | 0.577 | h1m | -0.449 |
| pmt | 0.528 | hnrnpub | -0.446 |
| thtpa | 0.521 | cd2bp2 | -0.429 |
| mycbp | 0.521 | nasp | -0.427 |
| gapdhs | 0.513 | stmn1a | -0.418 |
| hmgn3 | 0.512 | syncrip | -0.415 |
| gas6 | 0.512 | ppig | -0.405 |
| sparc | 0.498 | marcksb | -0.395 |
| dap1b | 0.490 | si:ch73-281n10.2 | -0.389 |
| anxa11b | 0.489 | akap12b | -0.386 |
| nck1a | 0.489 | anp32b | -0.384 |
| rnf213a | 0.486 | ncl | -0.382 |
| rnasekb | 0.486 | NC-002333.4 | -0.382 |
| cox18 | 0.482 | tbx16 | -0.378 |
| rps17 | 0.482 | hnrnpa1a | -0.375 |
| FQ976913.1 | 0.481 | tpx2 | -0.369 |
| itpr1b | 0.480 | ccna2 | -0.365 |
| selenou1a | 0.479 | hmgb2a | -0.364 |
| rps10 | 0.479 | hnrnpabb | -0.362 |
| med30 | 0.479 | npm1a | -0.359 |
| txndc5 | 0.477 | acin1b | -0.355 |
| pygl | 0.476 | hdgfl2 | -0.350 |
| tmem14ca | 0.476 | ebna1bp2 | -0.344 |
| fgf13a | 0.475 | baz1b | -0.343 |
| ndrg3b | 0.474 | gar1 | -0.342 |
| MCMDC2 | 0.473 | gra | -0.342 |
| lyst | 0.472 | ctcf | -0.342 |
| rpl28 | 0.471 | stm | -0.341 |
| vwc2 | 0.471 | ilf3b | -0.341 |
| gamt | 0.469 | pttg1 | -0.338 |
| zgc:92630 | 0.468 | srrm1 | -0.336 |
| mhc1zba | 0.468 | apoeb | -0.335 |
| gchfr | 0.467 | srrt | -0.333 |
| CT956002.2 | 0.467 | dnajc1 | -0.333 |