glutathione reductase
ZFIN 
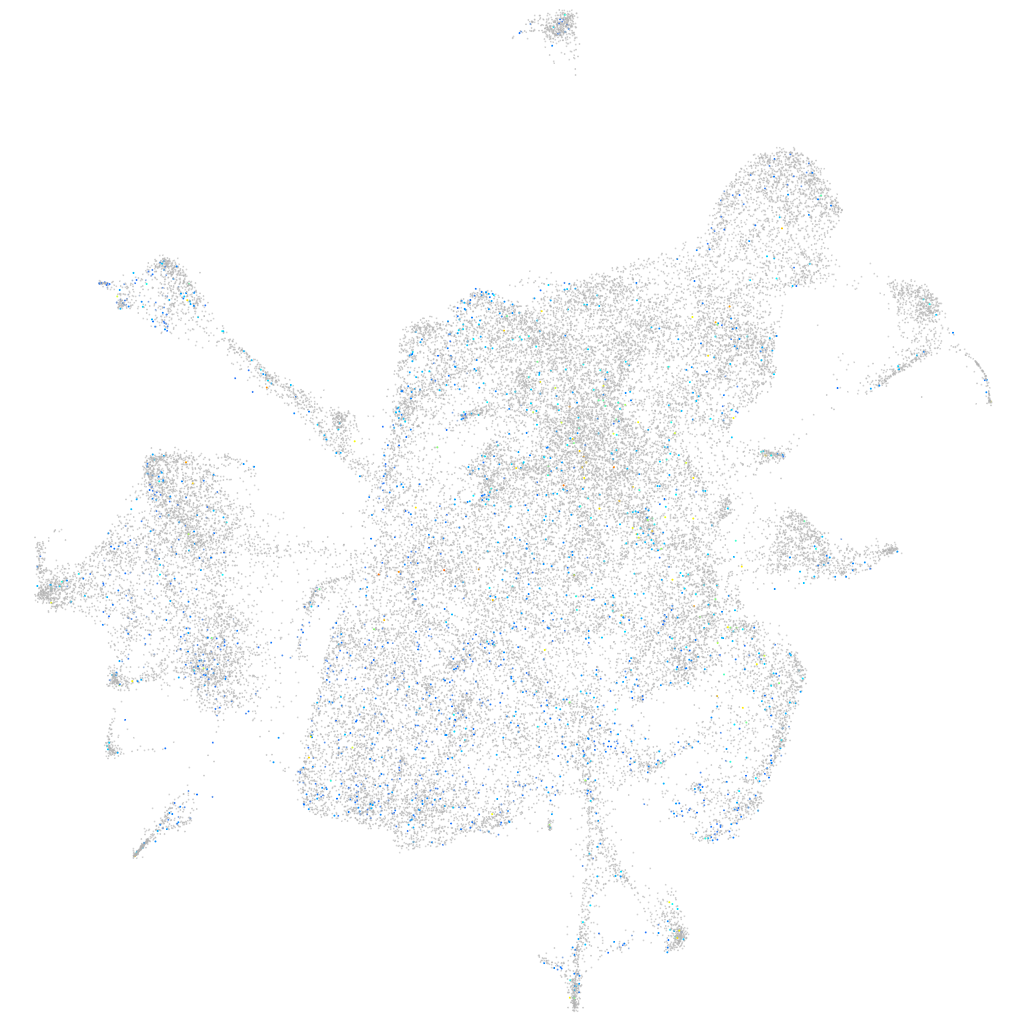
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| si:dkey-167k11.5 | 0.065 | fgfbp2b | -0.039 |
| si:dkey-148a17.5 | 0.064 | col2a1a | -0.038 |
| si:ch211-286b5.9 | 0.056 | col11a2 | -0.033 |
| si:ch211-189a21.1 | 0.052 | col9a2 | -0.033 |
| LOC101882775 | 0.052 | col9a1a | -0.033 |
| LOC110438615 | 0.052 | col9a3 | -0.032 |
| XLOC-039931 | 0.047 | col11a1a | -0.029 |
| CR318674.1 | 0.047 | cnmd | -0.028 |
| prdx1 | 0.045 | rps29 | -0.028 |
| si:dkey-73p2.1 | 0.044 | epyc | -0.028 |
| ubb | 0.044 | col2a1b | -0.028 |
| BX927261.1 | 0.043 | mia | -0.027 |
| zfp36l1a | 0.043 | matn1 | -0.027 |
| ap1s2 | 0.043 | matn4 | -0.025 |
| sept2 | 0.042 | ucmab | -0.025 |
| XLOC-019725 | 0.042 | acana | -0.024 |
| prdx2 | 0.042 | snorc | -0.024 |
| myl12.1 | 0.041 | pvalb2 | -0.022 |
| LOC101882203 | 0.041 | pvalb1 | -0.022 |
| mhc1lga | 0.040 | nme2b.2 | -0.022 |
| eef1a2 | 0.039 | wisp3 | -0.022 |
| pcbd1 | 0.039 | nog3 | -0.022 |
| gstp1 | 0.039 | VIT | -0.022 |
| tpm3 | 0.039 | tnxba | -0.021 |
| LOC100331720 | 0.039 | ptgdsa | -0.021 |
| LOC100538032.1 | 0.039 | otos | -0.020 |
| gng5 | 0.039 | actc1b | -0.020 |
| lsp1 | 0.039 | rpl38 | -0.019 |
| si:ch211-225b11.1 | 0.038 | wfdc2 | -0.019 |
| si:ch211-226h8.11 | 0.038 | ecrg4a | -0.019 |
| tubb4b | 0.038 | sox5 | -0.019 |
| bcam | 0.037 | itga10 | -0.019 |
| actb2 | 0.037 | si:dkey-6n6.1 | -0.018 |
| CR450814.1 | 0.037 | mylpfb | -0.018 |
| LOC101882072 | 0.037 | igsf10 | -0.018 |