gsg1-like
ZFIN 
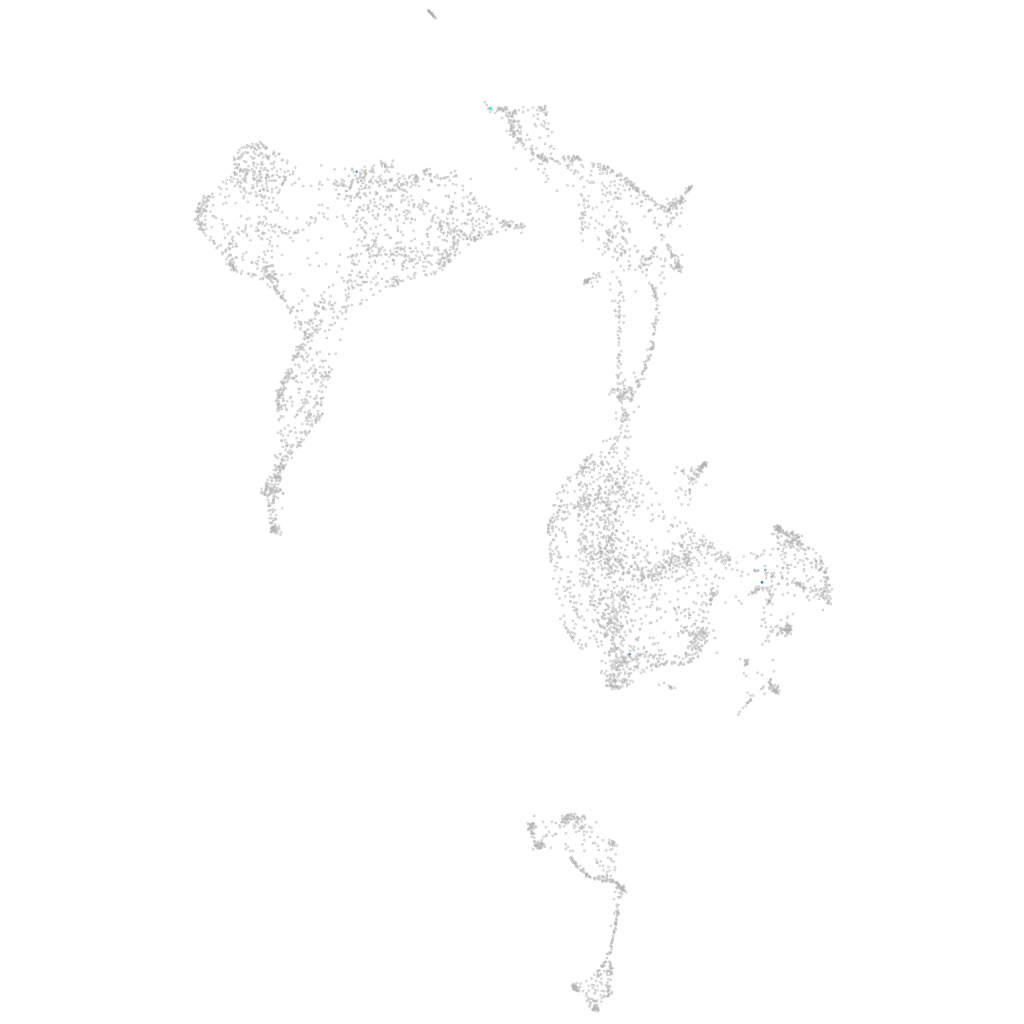
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| kif1c | 0.585 | tpt1 | -0.040 |
| lix1 | 0.365 | mt-co2 | -0.039 |
| pdgfc | 0.330 | rps18 | -0.029 |
| zgc:193726 | 0.327 | COX3 | -0.027 |
| gdf6b | 0.324 | myl12.1 | -0.026 |
| txlnbb | 0.318 | mt-co1 | -0.025 |
| cracr2ab | 0.276 | ppp2cb | -0.025 |
| tmem26a | 0.265 | rpl6 | -0.025 |
| trim35-27 | 0.260 | eif5a2 | -0.025 |
| si:ch1073-450f2.1 | 0.254 | fam32a | -0.024 |
| sim1a | 0.244 | rrbp1a | -0.024 |
| LOC103908938 | 0.233 | atp1a1a.1 | -0.024 |
| bmp6 | 0.218 | eif4g1a | -0.024 |
| tmem235 | 0.212 | mt-cyb | -0.023 |
| TTC28 | 0.210 | NC-002333.4 | -0.022 |
| si:ch211-176l24.4 | 0.206 | bsg | -0.022 |
| si:ch211-195h23.3 | 0.201 | rab5if | -0.022 |
| mif4gdb | 0.194 | kdelr2a | -0.022 |
| fgfrl1b | 0.193 | sec61a1 | -0.022 |
| sash3 | 0.183 | rpl5b | -0.022 |
| gtf2h1 | 0.176 | syncrip | -0.022 |
| ca10a | 0.167 | sar1b | -0.021 |
| brms1 | 0.165 | paip2b | -0.021 |
| tcp11l1 | 0.163 | srpra | -0.021 |
| BX323458.1 | 0.163 | tma7 | -0.021 |
| si:ch211-202h22.10 | 0.159 | oaz1a | -0.021 |
| ugt1b7 | 0.159 | mt-nd4 | -0.021 |
| v2rc1 | 0.159 | psmb4 | -0.021 |
| htr7a | 0.159 | eif3ha | -0.021 |
| LOC101882789 | 0.159 | srp72 | -0.020 |
| c16h2orf66 | 0.155 | zmat2 | -0.020 |
| metap2a | 0.155 | setb | -0.020 |
| fam151b | 0.152 | sec61a1l | -0.020 |
| CU019562.1 | 0.151 | sec63 | -0.020 |
| rprmb | 0.150 | calr3b | -0.020 |