glutaredoxin and cysteine rich domain containing 1 b
ZFIN 
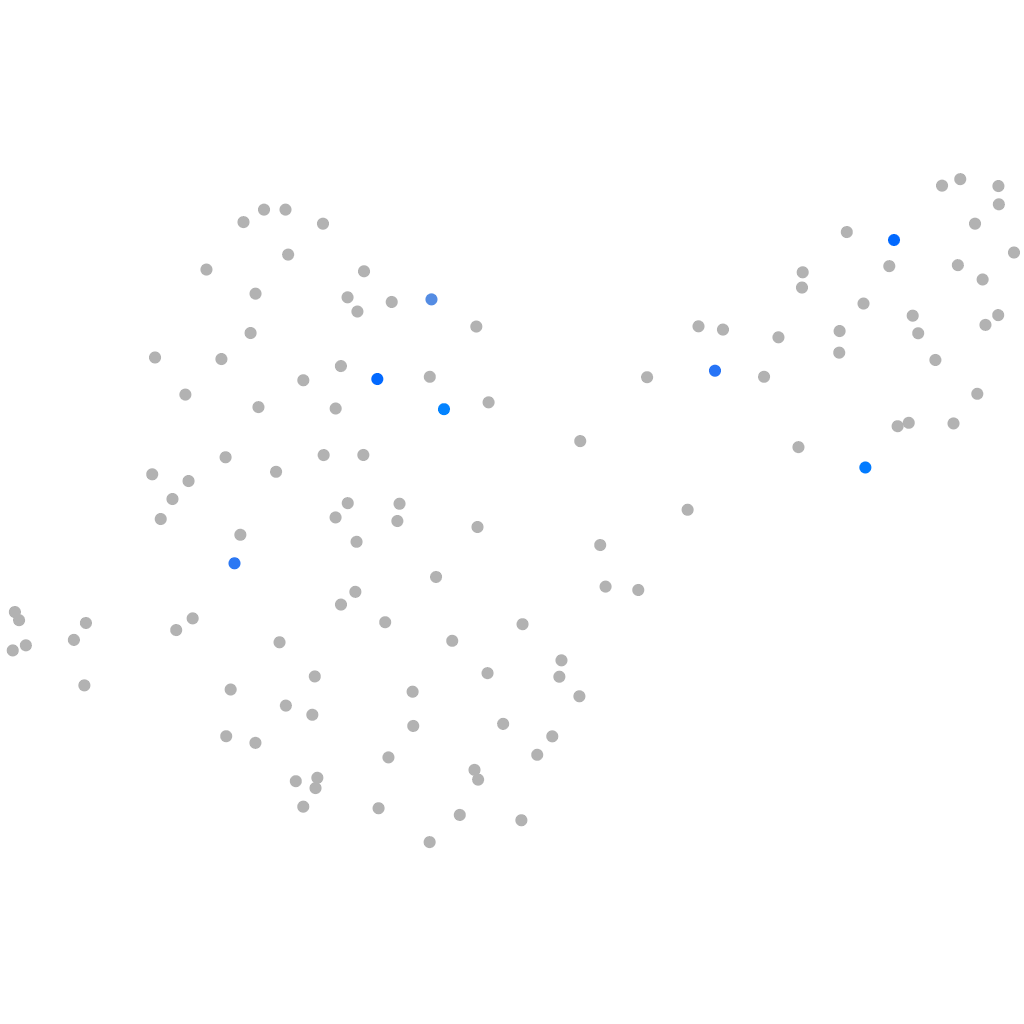
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 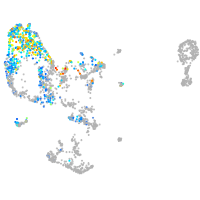 |
neural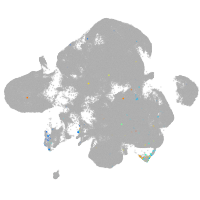 |
spinal cord / glia |
eye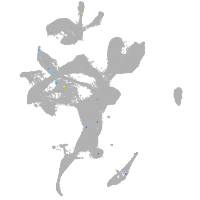 |
taste / olfactory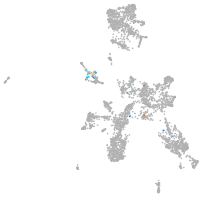 |
otic / lateral line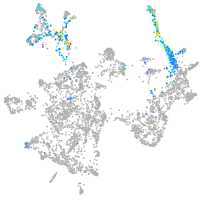 |
epidermis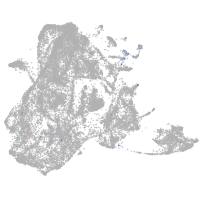 |
periderm |
fin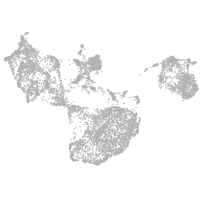 |
ionocytes / mucous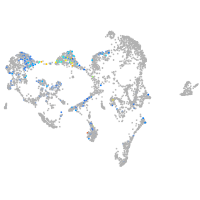 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dao.2 | 0.710 | hmga1a | -0.362 |
| trim35-33 | 0.698 | hnrnpaba | -0.345 |
| grinaa | 0.636 | hmgb2a | -0.341 |
| CR759843.2 | 0.629 | ncl | -0.327 |
| LOC108190785 | 0.614 | ccna2 | -0.315 |
| si:dkey-61p9.9 | 0.610 | dnajc1 | -0.283 |
| itga9 | 0.590 | hnrnpub | -0.281 |
| BX323559.1 | 0.590 | hnrnpabb | -0.281 |
| LOC101886429 | 0.590 | stm | -0.281 |
| si:dkey-148d16.5 | 0.588 | khdrbs1a | -0.280 |
| wnt4a | 0.588 | hnrnpa1b | -0.280 |
| CABZ01074130.1 | 0.588 | anp32e | -0.279 |
| ftr79 | 0.588 | h1m | -0.275 |
| si:dkey-9k7.3 | 0.572 | ptmab | -0.272 |
| fam8a1a | 0.564 | ctsla | -0.265 |
| CU638870.1 | 0.557 | ybx1 | -0.263 |
| MAJIN | 0.556 | actb1 | -0.259 |
| foxq2 | 0.552 | anp32a | -0.259 |
| LOC103911313 | 0.552 | NC-002333.4 | -0.258 |
| loxl2b | 0.552 | si:ch73-281n10.2 | -0.255 |
| si:ch211-198m1.1 | 0.552 | ilf3b | -0.252 |
| enpp6 | 0.551 | zc3h13 | -0.251 |
| tns1b | 0.550 | hnrnph1l | -0.249 |
| svopl | 0.549 | syncrip | -0.249 |
| LOC101882645 | 0.544 | hnrnpl2 | -0.248 |
| nlgn2b | 0.536 | tuba8l4 | -0.248 |
| tspeara | 0.536 | setb | -0.248 |
| BX511311.1 | 0.536 | hnrnpa1a | -0.241 |
| pm20d1.1 | 0.536 | stmn1a | -0.240 |
| stat4 | 0.535 | marcksb | -0.240 |
| si:dkey-34d22.3 | 0.527 | sde2 | -0.240 |
| pabpc4 | 0.527 | cactin | -0.235 |
| fcer1gl | 0.522 | srrt | -0.235 |
| enpep | 0.521 | snrpa | -0.235 |
| egln3 | 0.515 | hnrnpl | -0.234 |