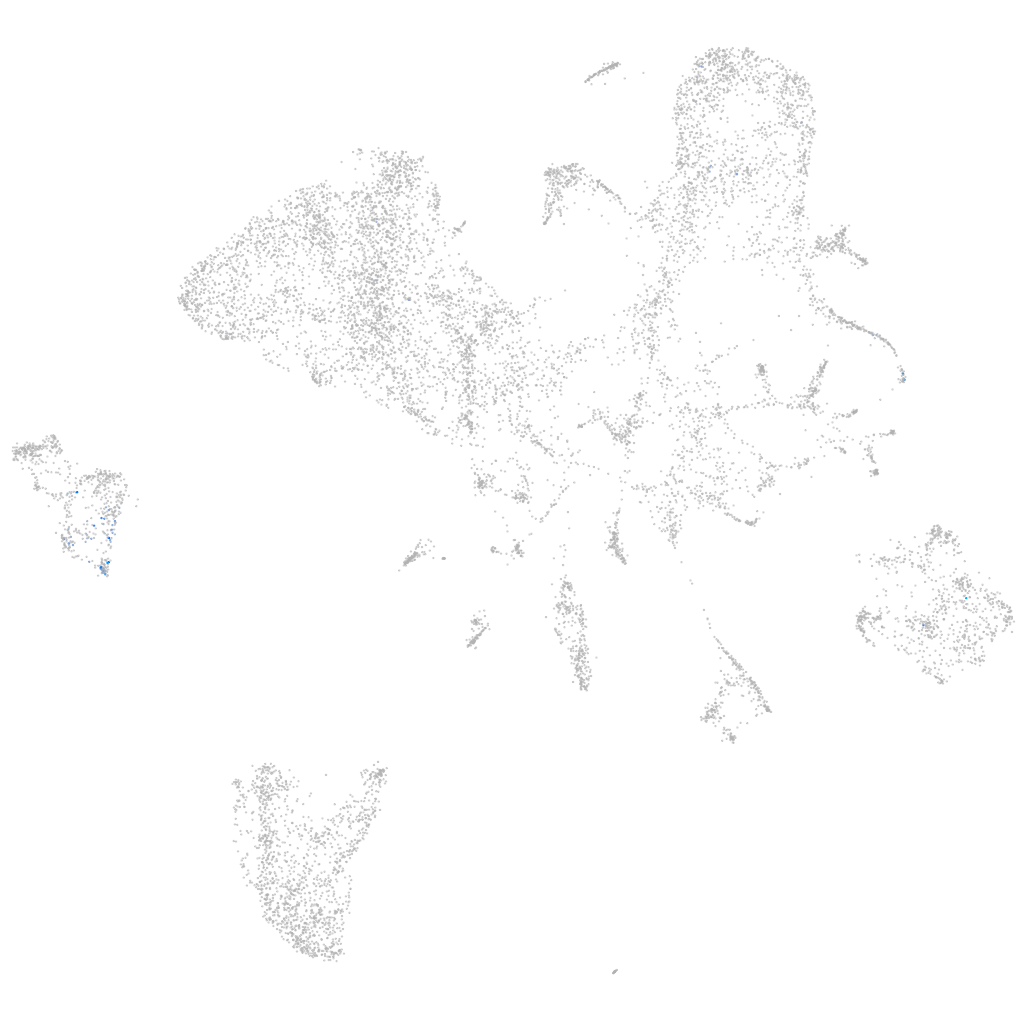
G protein-coupled receptor kinase 5
ZFIN 
Other cell groups


all cells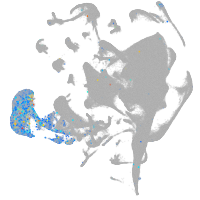 |
blastomeres |
PGCs |
endoderm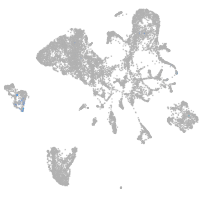 |
axial mesoderm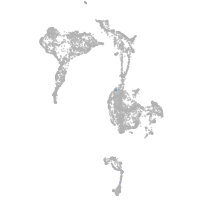 |
muscle 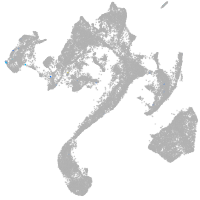 |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 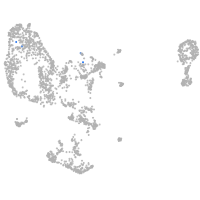 |
neural |
spinal cord / glia |
eye |
taste / olfactory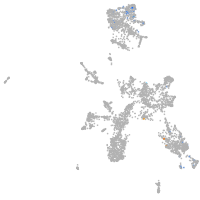 |
otic / lateral line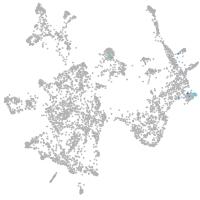 |
epidermis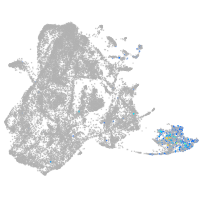 |
periderm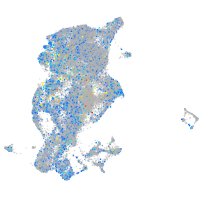 |
fin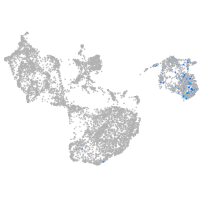 |
ionocytes / mucous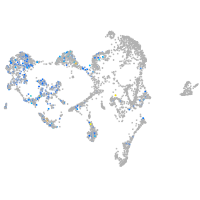 |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| CR926130.1 | 0.347 | gapdh | -0.064 |
| si:ch211-125o16.4 | 0.274 | gamt | -0.059 |
| krt1-19d | 0.273 | gatm | -0.052 |
| scel | 0.269 | bhmt | -0.052 |
| XLOC-002962 | 0.260 | fbp1b | -0.049 |
| si:ch211-157c3.4 | 0.254 | lgals2b | -0.047 |
| LOC100535170 | 0.252 | aqp12 | -0.047 |
| ANXA1 (1 of many) | 0.244 | gpx4a | -0.046 |
| evpla | 0.244 | mat1a | -0.046 |
| caspb | 0.242 | pklr | -0.045 |
| zgc:193505 | 0.241 | hspe1 | -0.045 |
| BX323854.1 | 0.241 | apoa4b.1 | -0.044 |
| LOC100149074 | 0.239 | grhprb | -0.044 |
| anxa1b | 0.238 | agxta | -0.044 |
| BX649431.2 | 0.236 | suclg1 | -0.043 |
| lye | 0.236 | cx32.3 | -0.043 |
| gna15.3 | 0.235 | scp2a | -0.043 |
| zgc:110333 | 0.233 | afp4 | -0.043 |
| cldne | 0.232 | apoc2 | -0.042 |
| sft2d2a | 0.225 | apoa1b | -0.042 |
| rab25a | 0.224 | pcbd1 | -0.041 |
| si:dkey-247k7.2 | 0.223 | kng1 | -0.041 |
| si:ch73-340m8.2 | 0.222 | dbi | -0.041 |
| sparta | 0.222 | agxtb | -0.041 |
| si:ch73-347e22.8 | 0.222 | apoa2 | -0.040 |
| anxa1c | 0.218 | ahcy | -0.040 |
| si:ch211-209f23.6 | 0.218 | BX908782.3 | -0.039 |
| gbgt1l3 | 0.217 | g6pca.2 | -0.039 |
| si:ch73-204p21.2 | 0.217 | tfa | -0.039 |
| gbgt1l4 | 0.216 | aldh7a1 | -0.039 |
| dhrs13a.2 | 0.215 | phyhd1 | -0.038 |
| ponzr2 | 0.215 | gcshb | -0.038 |
| si:dkeyp-97a10.1 | 0.213 | nipsnap3a | -0.038 |
| agr1 | 0.213 | gpd1b | -0.038 |
| zgc:100868 | 0.213 | pnp4b | -0.038 |