GRB2 related adaptor protein 2b
ZFIN 
Other cell groups


all cells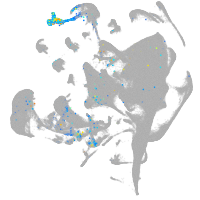 |
blastomeres |
PGCs |
endoderm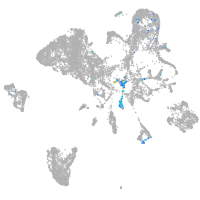 |
axial mesoderm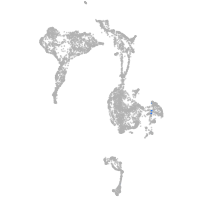 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme 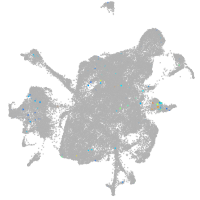 |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye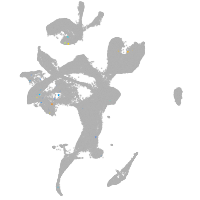 |
taste / olfactory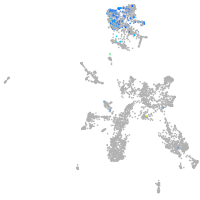 |
otic / lateral line |
epidermis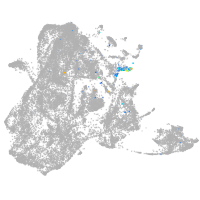 |
periderm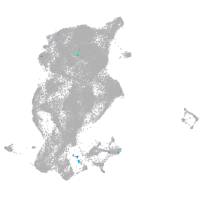 |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| coro1a | 0.700 | hbae3 | -0.360 |
| arpc1b | 0.634 | hbbe1.3 | -0.359 |
| rac2 | 0.633 | hbae1.3 | -0.347 |
| laptm5 | 0.630 | hbae1.1 | -0.346 |
| pfn1 | 0.625 | hbbe2 | -0.344 |
| cebpb | 0.591 | hbbe1.1 | -0.337 |
| ptpn6 | 0.557 | cahz | -0.320 |
| spi1b | 0.557 | hbbe1.2 | -0.306 |
| fam49ba | 0.557 | hemgn | -0.289 |
| ccr9a | 0.554 | alas2 | -0.279 |
| cotl1 | 0.552 | si:ch211-250g4.3 | -0.271 |
| arhgdig | 0.550 | fth1a | -0.270 |
| zgc:64051 | 0.546 | creg1 | -0.265 |
| wasb | 0.543 | zgc:56095 | -0.265 |
| glipr1a | 0.538 | nt5c2l1 | -0.258 |
| fcer1gl | 0.536 | blvrb | -0.257 |
| grap2a | 0.535 | slc4a1a | -0.249 |
| si:dkey-262k9.4 | 0.513 | zgc:163057 | -0.246 |
| ctss2.1 | 0.513 | lmo2 | -0.239 |
| wasa | 0.513 | epb41b | -0.235 |
| hcls1 | 0.512 | si:ch211-207c6.2 | -0.234 |
| si:ch211-102c2.4 | 0.504 | aqp1a.1 | -0.230 |
| cyba | 0.504 | tspo | -0.224 |
| eno3 | 0.499 | tmod4 | -0.220 |
| capgb | 0.494 | nmt1b | -0.211 |
| parvg | 0.483 | plac8l1 | -0.202 |
| FERMT3 (1 of many) | 0.481 | hbae5 | -0.198 |
| arpc4 | 0.479 | si:ch211-227m13.1 | -0.196 |
| ptprc | 0.477 | krt18a.1 | -0.191 |
| itgae.2 | 0.475 | sptb | -0.184 |
| rgs13 | 0.474 | hbbe3 | -0.184 |
| pycard | 0.472 | si:dkey-240n22.3 | -0.178 |
| arpc5b | 0.469 | dmtn | -0.177 |
| dub | 0.468 | uros | -0.176 |
| si:rp71-68n21.9 | 0.465 | tfr1a | -0.173 |




