granulito
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs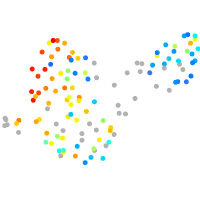 |
endoderm |
axial mesoderm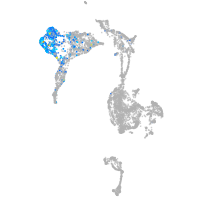 |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature 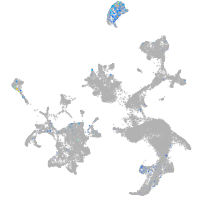 |
pronephros  |
neural |
spinal cord / glia |
eye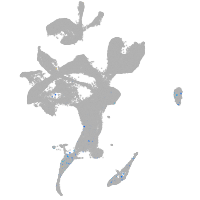 |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| dnd1 | 0.810 | rps27.1 | -0.705 |
| nasp | 0.759 | rpl37 | -0.675 |
| ddx4 | 0.731 | faua | -0.661 |
| ca15b | 0.723 | rps10 | -0.638 |
| h1m | 0.720 | zgc:114188 | -0.631 |
| marcksl1b | 0.714 | rps17 | -0.628 |
| stmn1a | 0.711 | rpl23 | -0.627 |
| surf2 | 0.708 | rps23 | -0.613 |
| tdrd7a | 0.705 | rnasekb | -0.610 |
| nanos3 | 0.669 | dap1b | -0.599 |
| stm | 0.669 | h3f3a | -0.595 |
| pttg1 | 0.662 | rpl13 | -0.594 |
| ppig | 0.644 | rps20 | -0.594 |
| fbl | 0.644 | rpl36a | -0.593 |
| akap12b | 0.640 | rps21 | -0.591 |
| zgc:77118 | 0.636 | cox8a | -0.586 |
| dkc1 | 0.630 | rps9 | -0.586 |
| cxcr4b | 0.627 | rps29 | -0.582 |
| NC-002333.4 | 0.622 | rplp0 | -0.582 |
| avd | 0.622 | rps14 | -0.580 |
| tbx16 | 0.621 | eef1db | -0.578 |
| tpx2 | 0.613 | si:dkey-151g10.6 | -0.577 |
| ddit4 | 0.612 | rpl22 | -0.573 |
| nop56 | 0.612 | rplp2 | -0.571 |
| sox11b | 0.607 | ppiab | -0.571 |
| hdgfl2 | 0.596 | rpl27a | -0.566 |
| gdf3 | 0.596 | ccni | -0.560 |
| hnrnpub | 0.595 | si:ch1073-429i10.3.1 | -0.560 |
| asb11 | 0.595 | rps19 | -0.556 |
| slbp | 0.595 | ldhba | -0.555 |
| bms1 | 0.586 | rpl5a | -0.555 |
| syncrip | 0.586 | rpl34 | -0.554 |
| slc16a3 | 0.583 | ISCU (1 of many) | -0.552 |
| ccna2 | 0.579 | cct2 | -0.547 |
| ncl | 0.576 | h3f3c | -0.542 |




