"glutamic pyruvate transaminase (alanine aminotransferase) 2, like"
ZFIN 
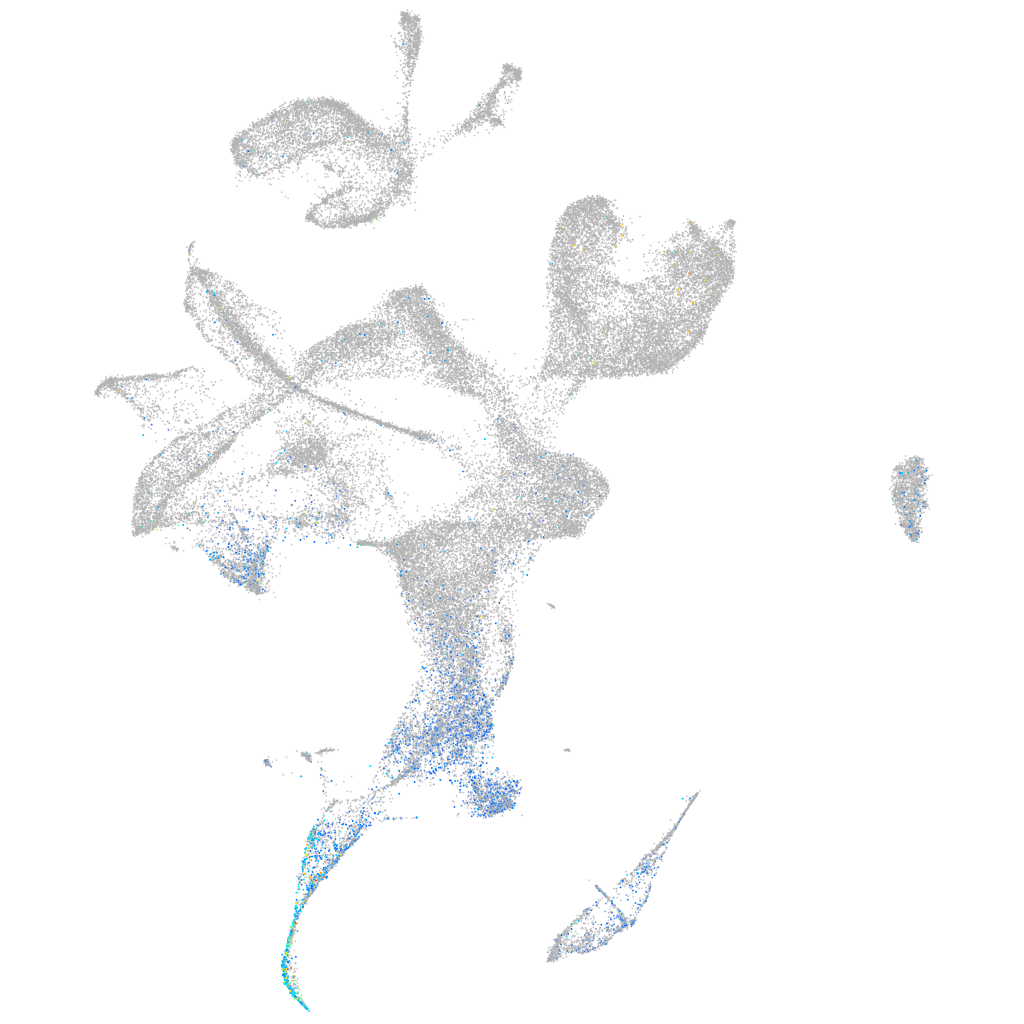
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| rgrb | 0.464 | ptmab | -0.267 |
| pmelb | 0.448 | si:ch73-1a9.3 | -0.206 |
| msnb | 0.445 | hmgn6 | -0.188 |
| selenou1b | 0.444 | ptmaa | -0.169 |
| tm6sf2 | 0.441 | gpm6aa | -0.157 |
| lratb.2 | 0.441 | tuba1c | -0.148 |
| dct | 0.440 | marcksl1b | -0.138 |
| fabp11b | 0.437 | hmgb1a | -0.131 |
| rpe65a | 0.435 | epb41a | -0.117 |
| tspan36 | 0.434 | nova2 | -0.112 |
| mb | 0.434 | h2afva | -0.111 |
| tyrp1b | 0.433 | snap25b | -0.111 |
| rlbp1b | 0.432 | hmgn2 | -0.108 |
| stra6 | 0.430 | tuba1a | -0.108 |
| pmela | 0.428 | cadm3 | -0.107 |
| pip4k2cb | 0.425 | hmgb3a | -0.107 |
| fgfbp1a | 0.424 | rtn1a | -0.107 |
| cracr2ab | 0.421 | anp32e | -0.106 |
| lrata | 0.420 | crx | -0.105 |
| gpr143 | 0.420 | neurod4 | -0.104 |
| pttg1ipb | 0.417 | stmn1b | -0.103 |
| tspan10 | 0.417 | foxg1b | -0.101 |
| rdh5 | 0.412 | gapdhs | -0.101 |
| bace2 | 0.409 | ndrg4 | -0.100 |
| rgra | 0.402 | si:ch1073-429i10.3.1 | -0.100 |
| oca2 | 0.398 | mdkb | -0.100 |
| slc7a2 | 0.398 | rorb | -0.099 |
| si:ch211-236l14.4 | 0.394 | atp1b2a | -0.099 |
| inpp5ka | 0.392 | h3f3c | -0.099 |
| inpp5kb | 0.390 | marcksl1a | -0.098 |
| sytl2a | 0.387 | ckbb | -0.097 |
| slc45a2 | 0.387 | hmgb1b | -0.097 |
| tyrp1a | 0.386 | mpp6b | -0.096 |
| rbp5 | 0.384 | fabp7a | -0.096 |
| slc16a1a | 0.383 | scrt2 | -0.094 |