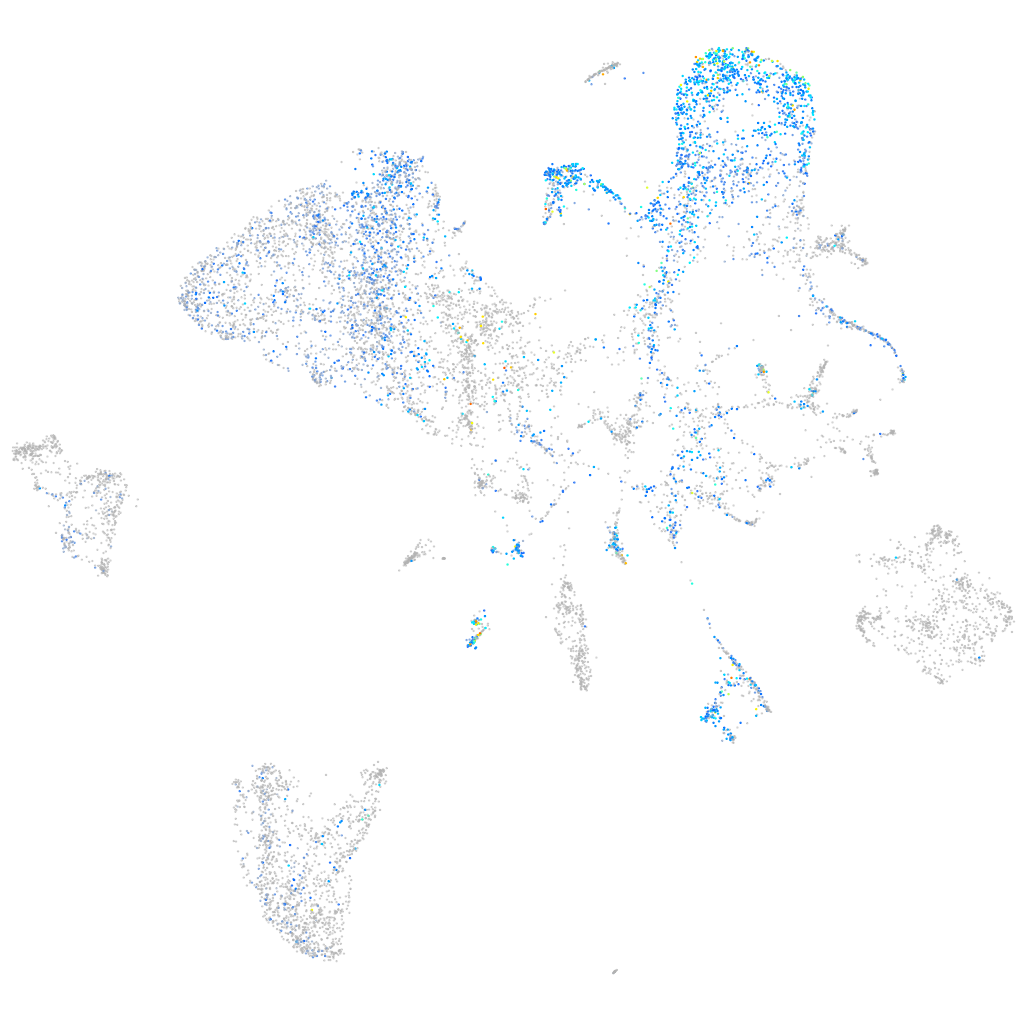
G protein-coupled receptor 39
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros  |
neural |
spinal cord / glia |
eye |
taste / olfactory |
otic / lateral line |
epidermis |
periderm |
fin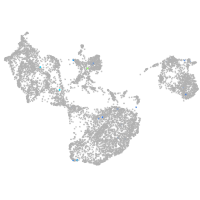 |
ionocytes / mucous |
pigment cells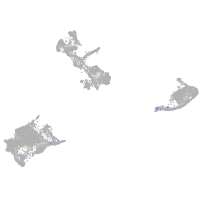 |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| chia.2 | 0.468 | aqp12 | -0.188 |
| slc15a1b | 0.460 | fabp3 | -0.186 |
| ace2 | 0.457 | bhmt | -0.176 |
| wu:fa56d06 | 0.455 | si:ch211-195b11.3 | -0.175 |
| cd36 | 0.454 | rtn1a | -0.166 |
| aqp8a.2 | 0.453 | prss1 | -0.154 |
| plac8.1 | 0.448 | prss59.2 | -0.154 |
| acsl5 | 0.444 | ela3l | -0.154 |
| rtn4b | 0.440 | ppdpfa | -0.153 |
| fabp2 | 0.439 | zgc:112160 | -0.153 |
| cpo | 0.439 | cpb1 | -0.152 |
| ada | 0.438 | erp27 | -0.151 |
| slc15a1a | 0.434 | CELA1 (1 of many) | -0.151 |
| gsta.1 | 0.432 | pdia2 | -0.151 |
| stoml3b | 0.431 | apoda.2 | -0.151 |
| zgc:158846 | 0.431 | ela2l | -0.150 |
| slc6a19a.2 | 0.429 | ctrb1 | -0.150 |
| slc5a1 | 0.425 | apoc1 | -0.149 |
| aoc1 | 0.425 | cpa5 | -0.149 |
| pdzk1 | 0.423 | fep15 | -0.149 |
| ldha | 0.423 | cel.1 | -0.149 |
| si:ch211-113d11.6 | 0.422 | sycn.2 | -0.148 |
| si:ch211-133l5.7 | 0.421 | prss59.1 | -0.148 |
| atp1a1a.4 | 0.416 | ela2 | -0.147 |
| pdzd3b | 0.413 | si:ch211-240l19.5 | -0.146 |
| s100a10a | 0.411 | cel.2 | -0.146 |
| dhrs1 | 0.410 | cpa4 | -0.145 |
| mbl2 | 0.408 | hspb1 | -0.145 |
| zgc:175280 | 0.407 | zgc:136461 | -0.145 |
| tmem86b | 0.406 | c6ast4 | -0.142 |
| crip1 | 0.406 | amy2a | -0.142 |
| rbp2a | 0.405 | zgc:153675 | -0.141 |
| mansc1 | 0.402 | endou | -0.139 |
| mogat2 | 0.402 | marcksl1b | -0.139 |
| si:ch211-161h7.8 | 0.402 | hmgb1b | -0.138 |