G protein-coupled receptor 37 like 1b
ZFIN 
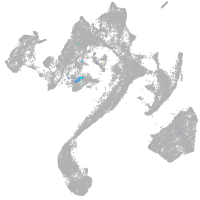
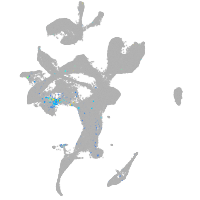
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc1a2b | 0.508 | hbbe3 | -0.038 |
| atp1a1b | 0.503 | lmo2 | -0.036 |
| slc6a11b | 0.480 | hhex | -0.029 |
| hepacama | 0.444 | dusp5 | -0.027 |
| atp1b4 | 0.426 | znfl2a | -0.027 |
| slc4a4a | 0.425 | f13a1b | -0.026 |
| slc1a3b | 0.409 | she | -0.026 |
| si:ch211-66e2.5 | 0.386 | cdh5 | -0.024 |
| zgc:165461 | 0.372 | si:ch211-156j16.1 | -0.024 |
| slc6a1b | 0.362 | urod | -0.024 |
| FO704813.1 | 0.358 | abracl | -0.024 |
| fabp7a | 0.337 | fli1b | -0.023 |
| gpm6bb | 0.327 | fli1a | -0.023 |
| cspg5b | 0.326 | etv2 | -0.023 |
| ppap2d | 0.324 | b3gnt3.4 | -0.023 |
| sept8b | 0.315 | blf | -0.023 |
| mlc1 | 0.310 | pfn1 | -0.023 |
| efhd1 | 0.303 | tmem88a | -0.023 |
| CU467822.1 | 0.293 | si:dkey-27i16.2 | -0.023 |
| mfge8a | 0.289 | gmfg | -0.022 |
| ptgdsb.2 | 0.289 | erg | -0.022 |
| eno1b | 0.288 | XLOC-014079 | -0.022 |
| CR848047.1 | 0.287 | XLOC-039757 | -0.022 |
| slc7a10b | 0.284 | clic2 | -0.022 |
| hspb15 | 0.284 | tal1 | -0.022 |
| cldn7a | 0.283 | cldng | -0.022 |
| acbd7 | 0.282 | alad | -0.022 |
| si:ch211-251b21.1 | 0.277 | capgb | -0.021 |
| slc7a10a | 0.274 | pecam1 | -0.021 |
| si:ch73-265d7.2 | 0.270 | yrk | -0.021 |
| nog2 | 0.260 | btk | -0.021 |
| mdkb | 0.259 | clec14a | -0.021 |
| endouc | 0.259 | klf3 | -0.021 |
| cdo1 | 0.252 | si:dkey-261j4.4 | -0.021 |
| fjx1 | 0.250 | si:dkeyp-97a10.2 | -0.021 |