G protein-coupled receptor 22a
ZFIN 
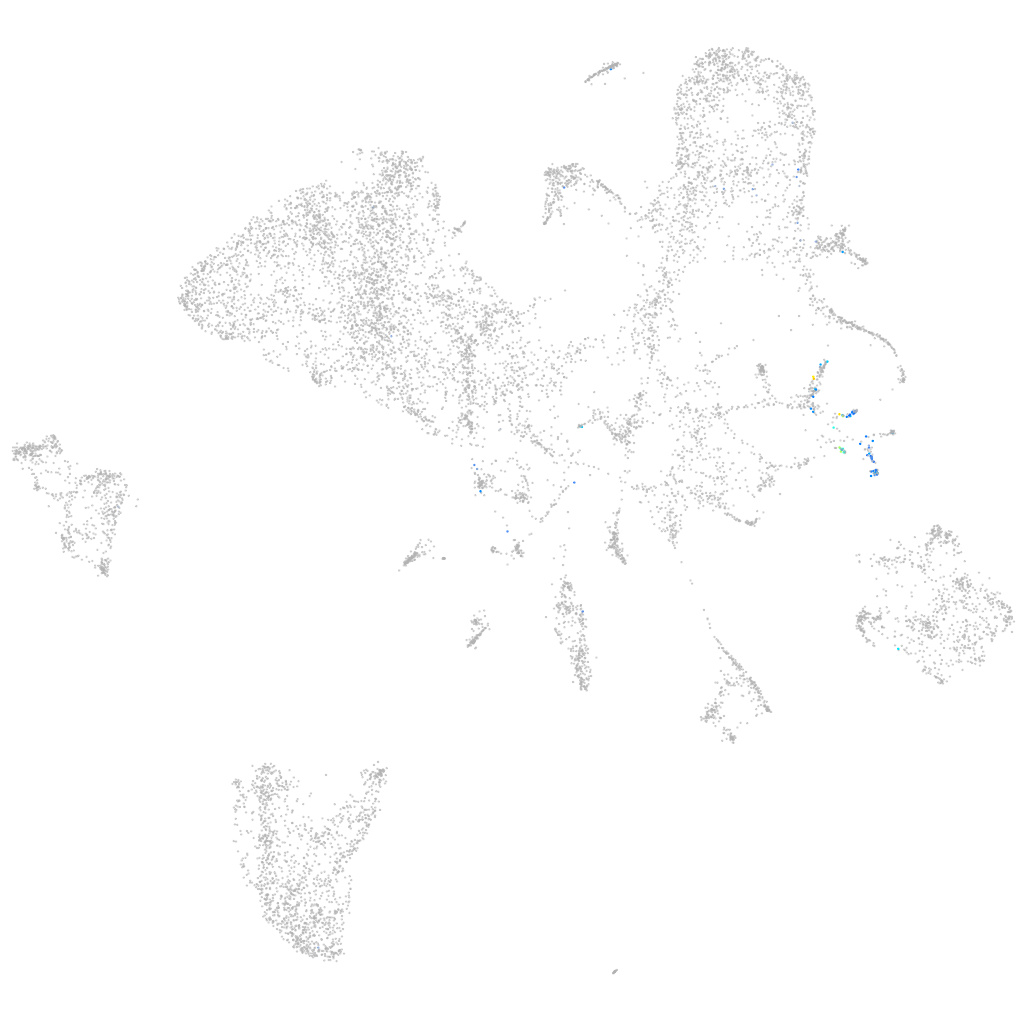
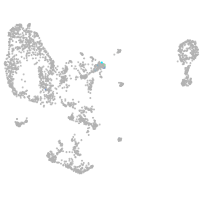
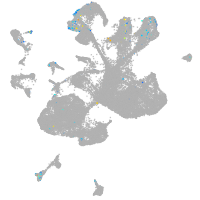
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| slc30a2 | 0.424 | ahcy | -0.089 |
| scgn | 0.414 | aldob | -0.086 |
| ndufa4l2a | 0.411 | gapdh | -0.083 |
| kcnj11 | 0.402 | rps20 | -0.080 |
| SLC5A10 | 0.385 | gstp1 | -0.075 |
| ins | 0.384 | gamt | -0.075 |
| si:dkey-153k10.9 | 0.371 | rplp2l | -0.073 |
| gcgb | 0.365 | eif4ebp1 | -0.073 |
| ptn | 0.363 | gstr | -0.070 |
| pcsk2 | 0.359 | gatm | -0.070 |
| isl1 | 0.352 | nupr1b | -0.068 |
| tspan7b | 0.339 | si:dkey-16p21.8 | -0.067 |
| rprmb | 0.331 | aldh6a1 | -0.066 |
| plppr5b | 0.327 | eno3 | -0.066 |
| syt1a | 0.322 | bhmt | -0.066 |
| pax6b | 0.317 | fbp1b | -0.065 |
| abhd15a | 0.309 | glud1b | -0.064 |
| rims2a | 0.309 | apoa1b | -0.063 |
| LOC100537384 | 0.302 | eef1da | -0.063 |
| trhra | 0.300 | gstt1a | -0.063 |
| scg3 | 0.300 | hdlbpa | -0.062 |
| kcnk9 | 0.299 | mat1a | -0.062 |
| rasd4 | 0.298 | aldh7a1 | -0.062 |
| CR774179.5 | 0.296 | cx32.3 | -0.060 |
| neurod1 | 0.296 | ppa1b | -0.060 |
| pcsk1nl | 0.294 | apoc2 | -0.060 |
| gcga | 0.291 | shmt1 | -0.060 |
| pnoca | 0.290 | fabp3 | -0.060 |
| scg2a | 0.281 | hspe1 | -0.060 |
| foxo1a | 0.280 | apoa4b.1 | -0.060 |
| dmtn | 0.279 | rpl11 | -0.059 |
| camk1da | 0.278 | afp4 | -0.058 |
| fam49bb | 0.278 | prdx6 | -0.058 |
| cacna1c | 0.274 | ckba | -0.058 |
| kcnma1a | 0.271 | ndufa4l | -0.058 |