G protein-coupled receptor 17
ZFIN 
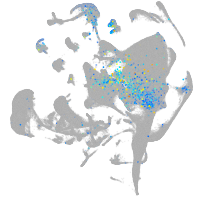
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| cntnap1 | 0.710 | rps27a | -0.040 |
| slitrk3a | 0.525 | rpl39 | -0.036 |
| si:ch73-335m24.2 | 0.481 | rpl7 | -0.033 |
| grm1a | 0.441 | rps25 | -0.032 |
| unm-hu7912 | 0.401 | rpl13a | -0.029 |
| aldoca | 0.378 | rpl17 | -0.029 |
| c7b | 0.352 | rps8a | -0.028 |
| tnr | 0.341 | rps3 | -0.025 |
| lingo1a | 0.329 | rack1 | -0.025 |
| lrp1aa | 0.321 | sec61g | -0.024 |
| cldn19 | 0.320 | eif2s2 | -0.023 |
| neto2a | 0.316 | rpl23a | -0.022 |
| pvalb7 | 0.306 | rps16 | -0.022 |
| kcnj11l | 0.303 | ssr1 | -0.022 |
| FP016056.1 | 0.294 | hspa8 | -0.021 |
| CR848841.1 | 0.294 | mt-co1 | -0.021 |
| plp1b | 0.287 | cox7b | -0.021 |
| si:ch211-108c17.2 | 0.275 | dap | -0.020 |
| CABZ01081748.1 | 0.258 | gatm | -0.019 |
| gdap1 | 0.257 | rps18 | -0.019 |
| tmeff2a | 0.252 | sec61b | -0.019 |
| lpcat2 | 0.247 | ssr2 | -0.019 |
| ptprdb | 0.245 | rps6 | -0.019 |
| nptnb | 0.244 | hdlbpa | -0.019 |
| CABZ01029822.1 | 0.243 | gapdh | -0.018 |
| XLOC-022150 | 0.235 | tmed10 | -0.018 |
| zgc:101853 | 0.234 | gamt | -0.018 |
| zgc:66433 | 0.226 | COX5B | -0.018 |
| pimr192 | 0.226 | rpl5a | -0.018 |
| eef1a1a | 0.226 | tram1 | -0.018 |
| pvalb6 | 0.223 | pomp | -0.017 |
| si:ch211-180a12.2 | 0.222 | cox6a1 | -0.017 |
| gpm6bb | 0.204 | aqp12 | -0.017 |
| ribc1 | 0.202 | rpl4 | -0.017 |
| tmem145 | 0.201 | hsp90b1 | -0.017 |