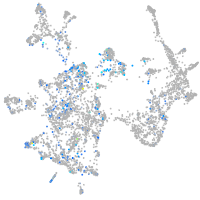
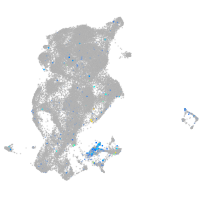
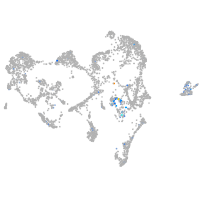
glycoprotein M6Bb
ZFIN 
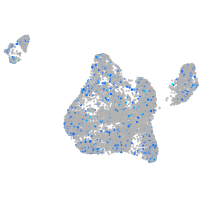
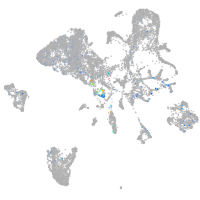
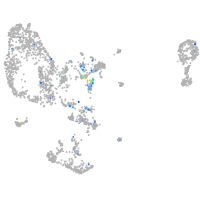
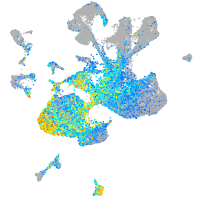
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| zgc:165461 | 0.343 | cox6c | -0.127 |
| CU467822.1 | 0.306 | cox7c | -0.126 |
| XLOC-003690 | 0.299 | cdh17 | -0.118 |
| mdka | 0.295 | eno3 | -0.114 |
| XLOC-003689 | 0.292 | mt-nd4 | -0.114 |
| CR751602.2 | 0.290 | cox6b2 | -0.114 |
| fabp7a | 0.287 | atp5po | -0.112 |
| hepacama | 0.282 | glud1b | -0.112 |
| gfap | 0.281 | cdaa | -0.109 |
| FP016213.1 | 0.279 | atp5if1a | -0.107 |
| her4.2 | 0.279 | aldh6a1 | -0.106 |
| cd82a | 0.276 | chchd10 | -0.106 |
| snai3 | 0.271 | cox5b2 | -0.106 |
| si:ch211-269k10.2 | 0.269 | bin2a | -0.104 |
| LOC110437771 | 0.266 | rplp2 | -0.103 |
| si:ch73-30b17.2 | 0.266 | upb1 | -0.102 |
| c1qc | 0.265 | sypl2a | -0.102 |
| si:ch211-66e2.5 | 0.262 | mt-nd5 | -0.101 |
| mdkb | 0.262 | mdh1aa | -0.101 |
| slitrk2 | 0.257 | elovl1b | -0.101 |
| sp8b | 0.256 | mt-co1 | -0.101 |
| CR848047.1 | 0.255 | mt-co2 | -0.100 |
| heyl | 0.255 | gapdh | -0.100 |
| si:ch211-251b21.1 | 0.254 | hoga1 | -0.100 |
| rfx4 | 0.250 | atp5f1d | -0.100 |
| slc1a3b | 0.245 | ndufb3 | -0.099 |
| gsx2 | 0.240 | sod2 | -0.099 |
| BX530079.1 | 0.240 | clic5b | -0.099 |
| wscd1b | 0.238 | cox7b | -0.098 |
| CU634008.1 | 0.238 | aldob | -0.098 |
| BX511023.1 | 0.236 | atp1a1a.4 | -0.098 |
| marcksl1a | 0.236 | mt-atp8 | -0.097 |
| LO018430.1 | 0.234 | sgk2a | -0.097 |
| zgc:153118 | 0.234 | cox6a2 | -0.097 |
| npas1 | 0.229 | atp1b1a | -0.096 |