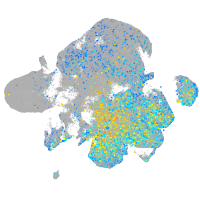
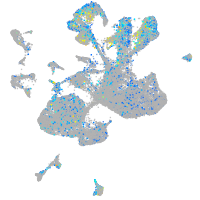
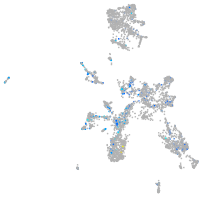
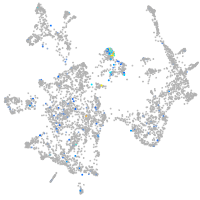
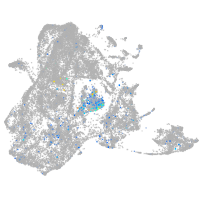
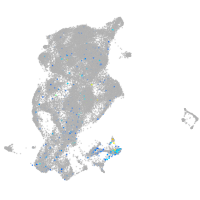
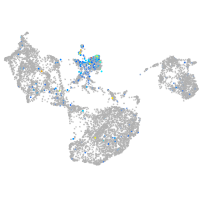
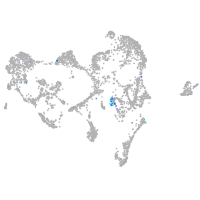
glycoprotein M6Ba
ZFIN 
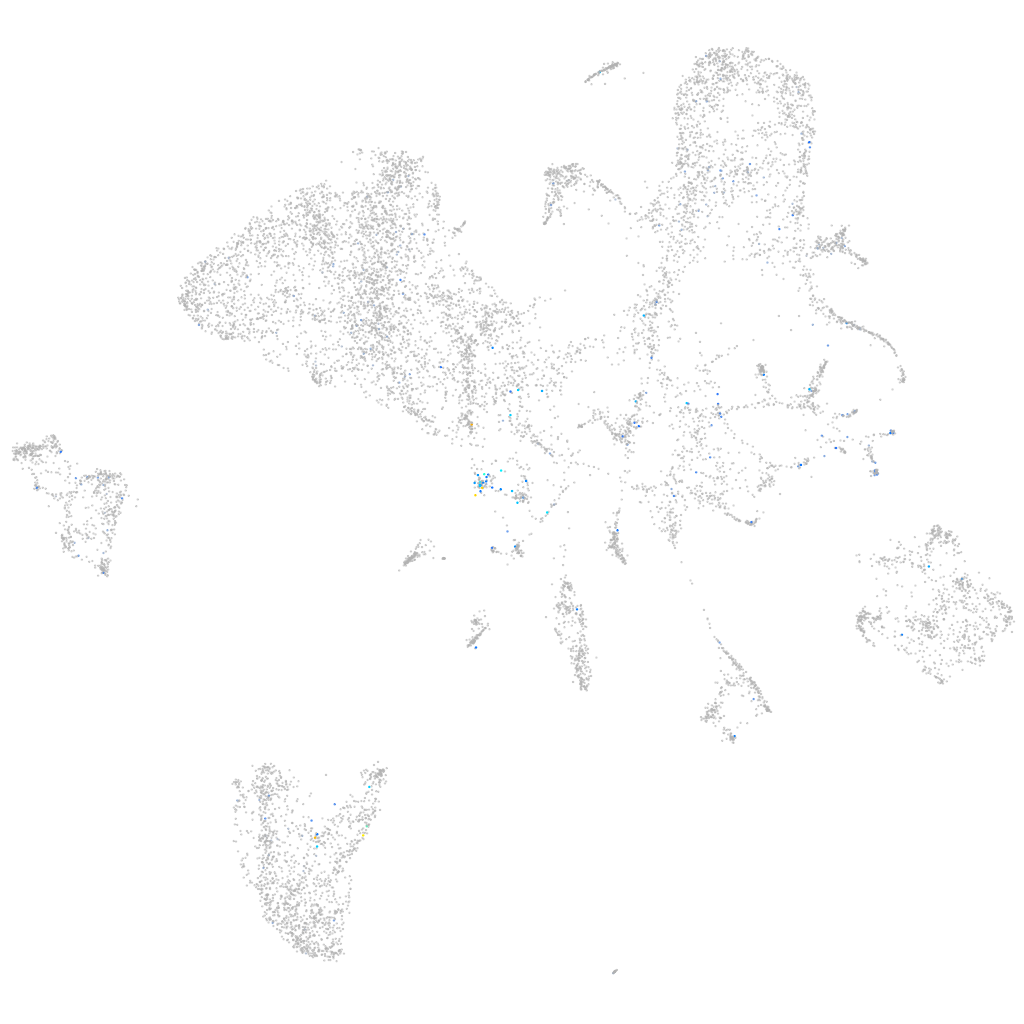
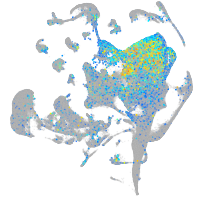
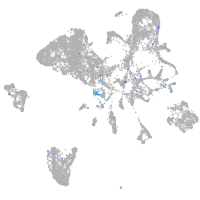
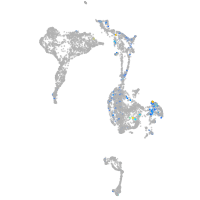
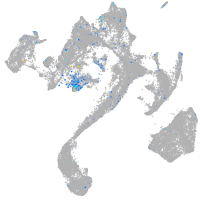
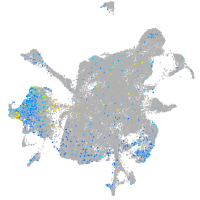
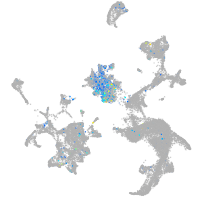
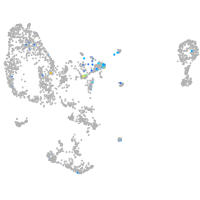
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| glrbb | 0.232 | rpl15 | -0.057 |
| XLOC-029934 | 0.226 | aldob | -0.056 |
| elavl4 | 0.225 | glud1b | -0.054 |
| XLOC-019901 | 0.223 | sec61b | -0.053 |
| ntrk3a | 0.222 | suclg1 | -0.051 |
| LO018020.2 | 0.219 | cox7a1 | -0.050 |
| wu:fj39g12 | 0.218 | fbp1b | -0.049 |
| lingo1a | 0.215 | nipsnap3a | -0.048 |
| apc2 | 0.212 | mgst1.2 | -0.048 |
| mmp24 | 0.210 | gnmt | -0.048 |
| rtn1b | 0.210 | scp2a | -0.047 |
| grapb | 0.207 | pgm1 | -0.047 |
| plppr4b | 0.207 | gapdh | -0.047 |
| gpm6aa | 0.206 | rdh1 | -0.046 |
| CABZ01046949.1 | 0.199 | pck2 | -0.046 |
| lrrn2 | 0.199 | gatm | -0.046 |
| pcdh1gc5 | 0.198 | cx32.3 | -0.045 |
| bin1a | 0.197 | gsta.1 | -0.045 |
| cnih2 | 0.195 | suclg2 | -0.045 |
| myt1la | 0.194 | cyp4v8 | -0.044 |
| fam189a1 | 0.193 | cyp3a65 | -0.044 |
| si:ch211-254n4.3 | 0.193 | eno3 | -0.044 |
| pimr94 | 0.192 | ckba | -0.044 |
| rab15 | 0.192 | gamt | -0.043 |
| pcdh1a | 0.191 | slc37a4a | -0.043 |
| nova2 | 0.189 | sdr16c5b | -0.043 |
| gabrb4 | 0.189 | acadm | -0.043 |
| elavl3 | 0.189 | dcxr | -0.043 |
| caly | 0.186 | cyp2n13 | -0.043 |
| sv2a | 0.185 | atox1 | -0.042 |
| LOC110437922 | 0.183 | f2 | -0.042 |
| si:dkeyp-74b6.2 | 0.183 | gpx4a | -0.042 |
| neurod2 | 0.182 | ppa1b | -0.042 |
| stxbp1a | 0.182 | vmo1b | -0.042 |
| scdb | 0.178 | shmt1 | -0.042 |