gon-4-like (C. elegans)
ZFIN 
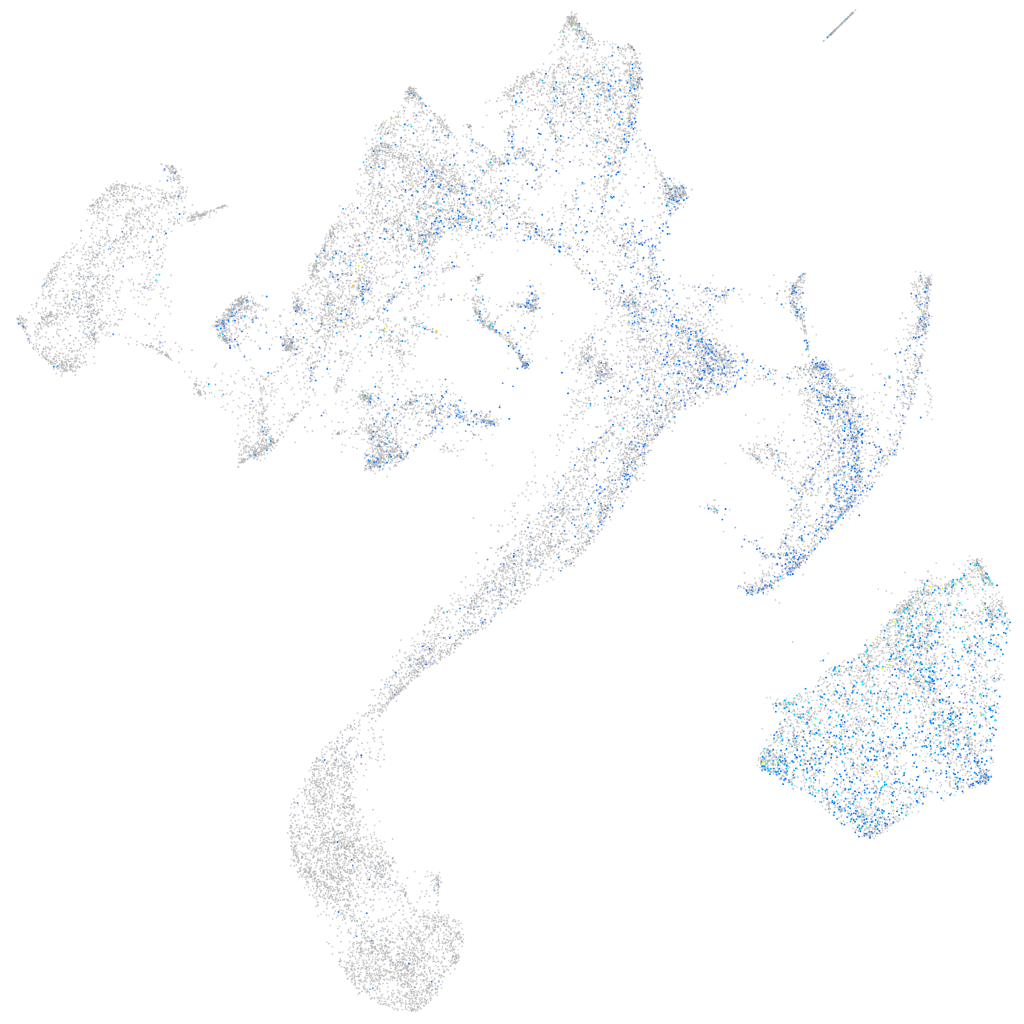
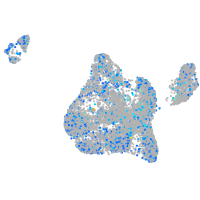
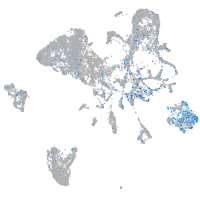
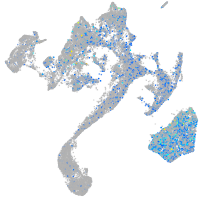
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| hnrnpub | 0.240 | rpl37 | -0.215 |
| hnrnpa1b | 0.239 | zgc:114188 | -0.203 |
| acin1a | 0.234 | rps10 | -0.200 |
| seta | 0.233 | rps17 | -0.191 |
| nucks1a | 0.229 | actc1b | -0.188 |
| anp32e | 0.227 | vdac3 | -0.167 |
| smc1al | 0.226 | ak1 | -0.164 |
| marcksb | 0.226 | ckma | -0.163 |
| lig1 | 0.225 | ckmb | -0.162 |
| NC-002333.4 | 0.225 | atp2a1 | -0.159 |
| hnrnpm | 0.225 | aldoab | -0.158 |
| mki67 | 0.225 | ttn.2 | -0.158 |
| cx43.4 | 0.224 | atp5meb | -0.157 |
| rbm4.3 | 0.224 | vdac2 | -0.157 |
| ilf3b | 0.223 | fabp3 | -0.156 |
| cdx4 | 0.222 | tnnc2 | -0.154 |
| hnrnpa1a | 0.222 | tmem38a | -0.153 |
| snrnp70 | 0.221 | mylpfa | -0.151 |
| anp32a | 0.221 | pabpc4 | -0.151 |
| brd3a | 0.220 | ttn.1 | -0.151 |
| cirbpa | 0.220 | gamt | -0.148 |
| ppig | 0.219 | suclg1 | -0.148 |
| ppm1g | 0.219 | mdh2 | -0.147 |
| safb | 0.219 | tpma | -0.147 |
| hmga1a | 0.219 | eef1da | -0.147 |
| anp32b | 0.218 | atp5if1b | -0.147 |
| ncl | 0.217 | acta1b | -0.147 |
| srsf1a | 0.217 | neb | -0.146 |
| chaf1a | 0.217 | mybphb | -0.145 |
| top1l | 0.217 | atp5mc3b | -0.145 |
| banf1 | 0.213 | eno1a | -0.144 |
| dek | 0.212 | CABZ01078594.1 | -0.144 |
| hnrnpabb | 0.211 | eno3 | -0.144 |
| syncrip | 0.211 | tpi1b | -0.143 |
| srrm1 | 0.211 | romo1 | -0.143 |