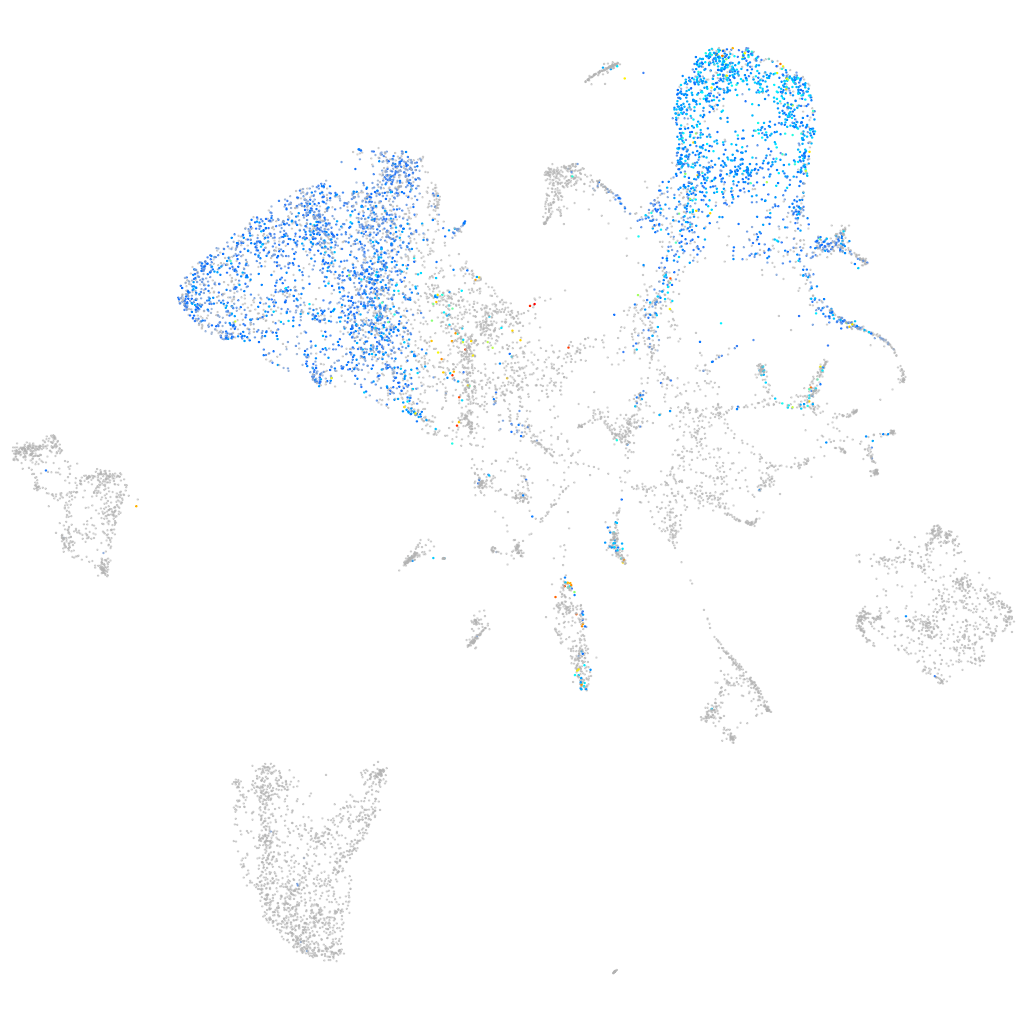
golgi transport 1A
ZFIN 
Other cell groups


all cells |
blastomeres |
PGCs |
endoderm |
axial mesoderm |
muscle  |
mural cells / non-skeletal muscle  |
mesenchyme  |
hematopoietic / vasculature  |
pronephros 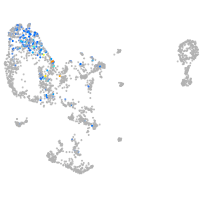 |
neural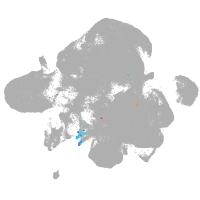 |
spinal cord / glia |
eye |
taste / olfactory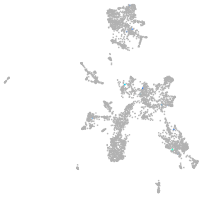 |
otic / lateral line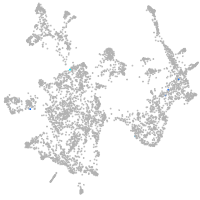 |
epidermis |
periderm |
fin |
ionocytes / mucous |
pigment cells |
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| ugt1a7 | 0.548 | slc38a5b | -0.265 |
| rbp2a | 0.521 | rtn1a | -0.236 |
| si:ch211-107o10.3 | 0.511 | si:ch211-195b11.3 | -0.232 |
| ada | 0.508 | ctrl | -0.218 |
| sult2st2 | 0.505 | hmgb3a | -0.217 |
| mttp | 0.502 | epcam | -0.215 |
| srd5a2a | 0.501 | glulb | -0.214 |
| ace2 | 0.500 | marcksb | -0.211 |
| aqp8a.2 | 0.500 | jpt1b | -0.208 |
| dhrs1 | 0.496 | marcksl1b | -0.208 |
| cd36 | 0.493 | qkia | -0.205 |
| dhrs9 | 0.490 | cpb1 | -0.205 |
| rdh1 | 0.486 | zgc:112160 | -0.205 |
| glud1b | 0.484 | fep15 | -0.204 |
| chia.2 | 0.483 | apoda.2 | -0.204 |
| mogat2 | 0.479 | id1 | -0.203 |
| fabp2 | 0.478 | cldnb | -0.203 |
| cyp8b1 | 0.477 | prss59.1 | -0.203 |
| apoc2 | 0.477 | cpa5 | -0.203 |
| wu:fa56d06 | 0.474 | ela2l | -0.202 |
| agmo | 0.474 | pdia2 | -0.202 |
| si:ch211-161h7.8 | 0.473 | erp27 | -0.202 |
| zgc:153968 | 0.472 | sycn.2 | -0.202 |
| apoa4b.1 | 0.472 | prss59.2 | -0.201 |
| si:ch211-133l5.7 | 0.470 | ela3l | -0.201 |
| gcshb | 0.468 | CELA1 (1 of many) | -0.200 |
| abcc2 | 0.468 | si:ch211-240l19.5 | -0.200 |
| lta4h | 0.468 | ela2 | -0.200 |
| gpx4a | 0.468 | hmgb1b | -0.200 |
| sdr16c5b | 0.467 | ctrb1 | -0.200 |
| cyp2y3 | 0.465 | cel.1 | -0.199 |
| slc6a19a.2 | 0.465 | cx43.4 | -0.198 |
| tmem86b | 0.465 | cel.2 | -0.198 |
| si:dkey-36i7.3 | 0.464 | cldn7b | -0.197 |
| fdx1 | 0.463 | cd81a | -0.196 |