N-acetylglucosamine-1-phosphate transferase subunit gamma
ZFIN 
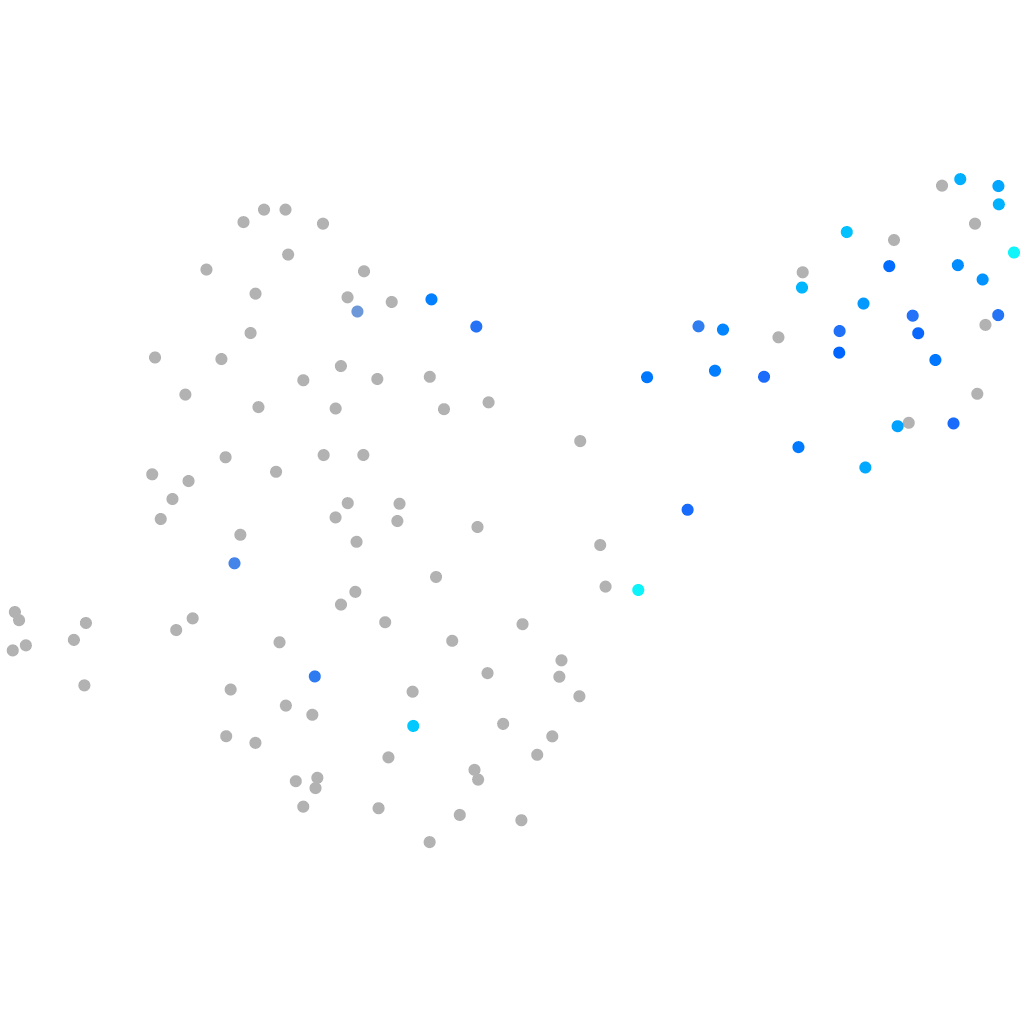
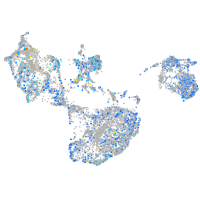
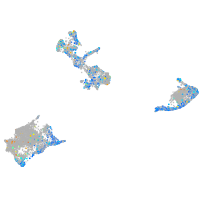
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| AL935186.9 | 0.673 | NC-002333.4 | -0.538 |
| zgc:92744 | 0.667 | brd3a | -0.498 |
| rps17 | 0.654 | hnrnpa1a | -0.494 |
| ewsr1b | 0.654 | ncl | -0.479 |
| sinhcafl | 0.643 | hnrnpa1b | -0.478 |
| cct2 | 0.629 | ppig | -0.469 |
| ISCU (1 of many) | 0.624 | npm1a | -0.467 |
| derl2 | 0.622 | acin1b | -0.463 |
| tnrc6a | 0.619 | cdk11b | -0.463 |
| zgc:56493 | 0.618 | fbl | -0.457 |
| rps10 | 0.615 | srrm1 | -0.446 |
| mvb12a | 0.611 | top1l | -0.445 |
| hnrnpa0l | 0.591 | akap12b | -0.440 |
| tma16 | 0.590 | marcksb | -0.437 |
| exosc3 | 0.580 | marcksl1b | -0.436 |
| rpl37 | 0.575 | tpx2 | -0.432 |
| dap1b | 0.572 | nop58 | -0.432 |
| MCMDC2 | 0.570 | nucks1a | -0.415 |
| zgc:86839 | 0.567 | nop56 | -0.406 |
| zgc:171566 | 0.562 | tbx16 | -0.406 |
| ndufa3 | 0.559 | ISCU | -0.401 |
| gsta.1 | 0.558 | stm | -0.398 |
| marcksl1a | 0.557 | hnrnpub | -0.397 |
| hint1 | 0.557 | baz1b | -0.395 |
| zgc:114188 | 0.556 | srrt | -0.395 |
| prdx4 | 0.555 | apoeb | -0.391 |
| calm3a | 0.552 | htatsf1 | -0.391 |
| tegt | 0.551 | gar1 | -0.389 |
| ghra | 0.550 | ebna1bp2 | -0.389 |
| rsrc2 | 0.548 | snrpb2 | -0.387 |
| COX5B | 0.547 | ddx18 | -0.384 |
| ndufb3 | 0.545 | mphosph10 | -0.379 |
| esd | 0.543 | snrpa | -0.379 |
| selenom | 0.542 | h1m | -0.375 |
| rnasekb | 0.541 | elavl1a | -0.375 |