glycine N-methyltransferase
ZFIN 
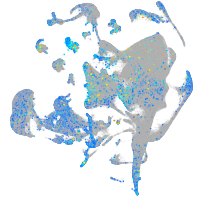
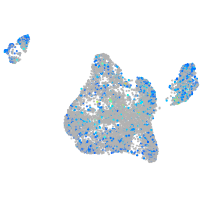
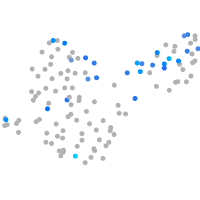
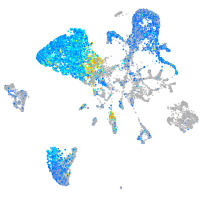
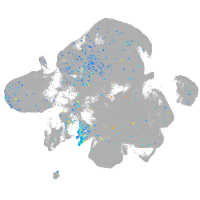
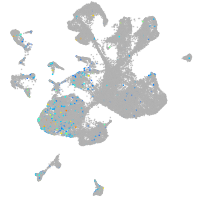
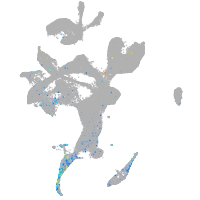
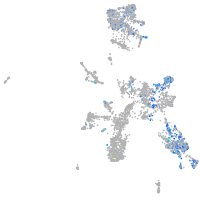
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| prdx1 | 0.215 | ptmaa | -0.091 |
| sprb | 0.191 | mdkb | -0.084 |
| pts | 0.183 | tuba1c | -0.069 |
| akr1b1 | 0.182 | gpm6aa | -0.069 |
| cyb5a | 0.182 | stmn1b | -0.066 |
| cx30.3 | 0.175 | tyrp1b | -0.066 |
| gch2 | 0.171 | chd4a | -0.065 |
| gpd1b | 0.167 | nova2 | -0.064 |
| impdh1b | 0.163 | ptmab | -0.064 |
| paics | 0.158 | dct | -0.062 |
| mibp | 0.158 | tyrp1a | -0.060 |
| atic | 0.156 | icn | -0.059 |
| si:dkey-251i10.2 | 0.151 | sox11b | -0.057 |
| aox5 | 0.150 | si:ch211-222l21.1 | -0.056 |
| ppat | 0.149 | zgc:158463 | -0.055 |
| oacyl | 0.148 | sox4a | -0.055 |
| slc2a11b | 0.147 | LOC103909099 | -0.055 |
| rbp4l | 0.147 | zc4h2 | -0.055 |
| tmem130 | 0.145 | rnasekb | -0.054 |
| CABZ01021592.1 | 0.145 | hbae3 | -0.054 |
| iah1 | 0.144 | defbl1 | -0.054 |
| bco1 | 0.144 | foxp4 | -0.054 |
| glulb | 0.143 | tuba1a | -0.053 |
| prtfdc1 | 0.140 | si:ch211-105c13.3 | -0.052 |
| uraha | 0.140 | elavl3 | -0.052 |
| CABZ01032488.1 | 0.139 | oca2 | -0.052 |
| TMEM19 | 0.138 | apoda.1 | -0.052 |
| kctd12.2 | 0.137 | zgc:91968 | -0.052 |
| cyb561a3b | 0.136 | pmela | -0.052 |
| slc22a7a | 0.136 | hmgb1b | -0.051 |
| prps1a | 0.132 | hmgb1a | -0.051 |
| cax1 | 0.130 | nptna | -0.051 |
| sigmar1 | 0.129 | sncb | -0.051 |
| rgs2 | 0.128 | tmeff1b | -0.051 |
| sult1st1 | 0.127 | FO082781.1 | -0.050 |