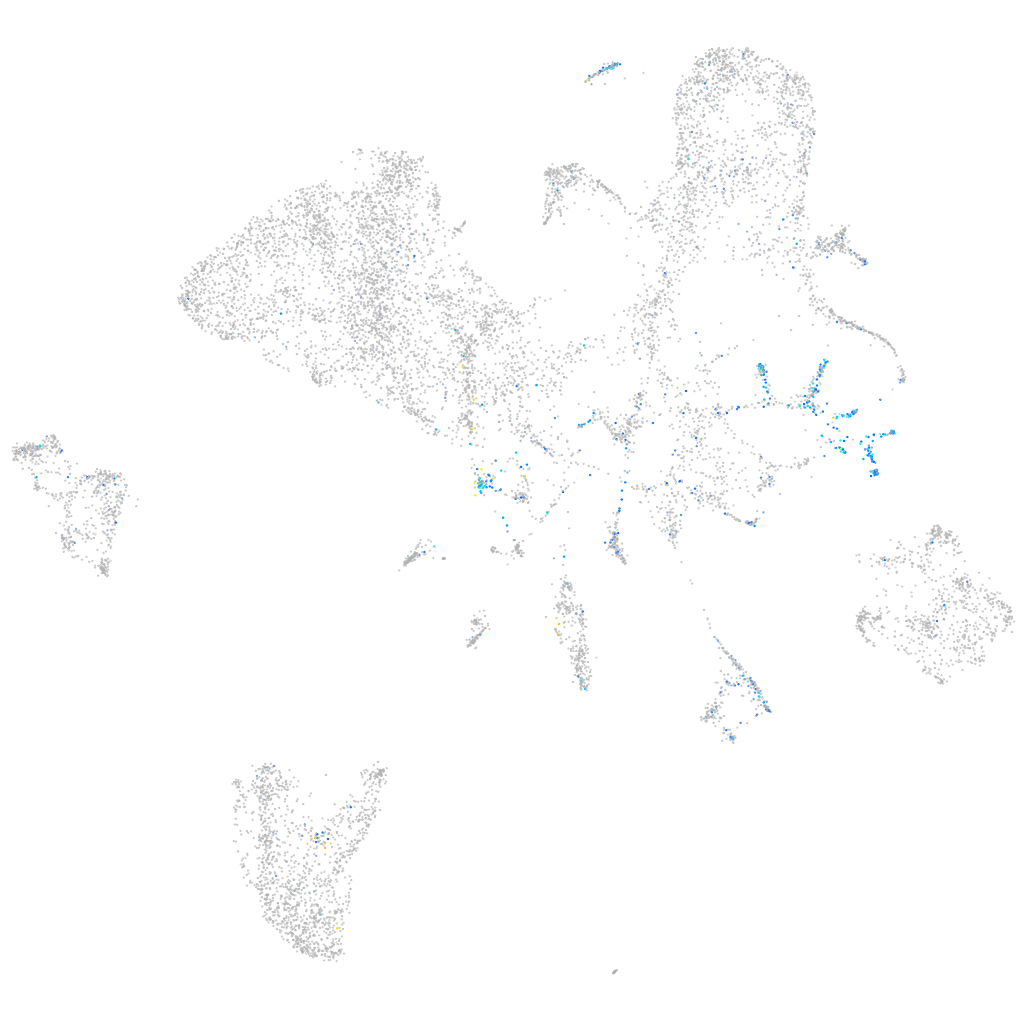
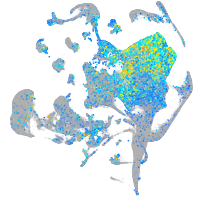
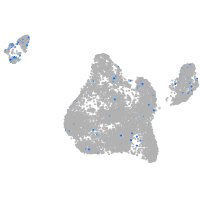
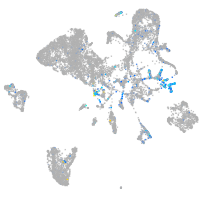
"guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a"
ZFIN 
Expression by stage/cluster


Correlated gene expression


| Positive correlation | Negative correlation | ||
| Gene | r | Gene | r |
| neurod1 | 0.396 | gapdh | -0.168 |
| scg3 | 0.389 | aldob | -0.166 |
| syt1a | 0.377 | ahcy | -0.163 |
| pax6b | 0.376 | gamt | -0.154 |
| vamp2 | 0.370 | hdlbpa | -0.143 |
| scgn | 0.369 | mat1a | -0.141 |
| vat1 | 0.363 | gatm | -0.139 |
| kcnj11 | 0.352 | fbp1b | -0.137 |
| rnasekb | 0.349 | aldh7a1 | -0.136 |
| pcsk1nl | 0.349 | aldh6a1 | -0.135 |
| gng3 | 0.343 | si:dkey-16p21.8 | -0.134 |
| stxbp1a | 0.343 | gstr | -0.133 |
| insm1a | 0.336 | eno3 | -0.129 |
| atp6v0cb | 0.334 | mgst1.2 | -0.129 |
| si:dkey-153k10.9 | 0.334 | agxtb | -0.127 |
| sncb | 0.328 | gstt1a | -0.126 |
| scg2a | 0.327 | cx32.3 | -0.126 |
| tuba1c | 0.324 | agxta | -0.125 |
| mir7a-1 | 0.324 | nupr1b | -0.125 |
| pcsk1 | 0.323 | apoa4b.1 | -0.125 |
| sypa | 0.322 | glud1b | -0.123 |
| atpv0e2 | 0.321 | pnp4b | -0.119 |
| elavl4 | 0.321 | bhmt | -0.119 |
| id4 | 0.319 | apoa1b | -0.118 |
| myt1b | 0.319 | scp2a | -0.118 |
| tspan7b | 0.317 | aqp12 | -0.117 |
| isl1 | 0.315 | gnmt | -0.117 |
| rims2a | 0.315 | suclg2 | -0.117 |
| tmsb | 0.315 | tdo2a | -0.117 |
| mllt11 | 0.314 | nipsnap3a | -0.116 |
| cnrip1a | 0.314 | ugt1a7 | -0.116 |
| atp1a3a | 0.312 | apoc2 | -0.116 |
| aldocb | 0.308 | isoc2 | -0.115 |
| rprmb | 0.307 | g6pca.2 | -0.113 |
| sv2a | 0.305 | apom | -0.113 |